Research Article
Volume 1 Issue 2 - 2017
Role of Quantum Chemical Parameters: Modeling of HIV-1 Inhibition Activity for CCR5 Antagonists
1Department of Chemistry, Jain University, Bangalore, Karnataka, INDIA
2Department of Chemistry, Symbiosis University of Applied Sciences, Indore, INDIA
3Department of Applied Sciences, National Institute of Technical Teachers Training and Research, Bhopal, INDIA
2Department of Chemistry, Symbiosis University of Applied Sciences, Indore, INDIA
3Department of Applied Sciences, National Institute of Technical Teachers Training and Research, Bhopal, INDIA
*Corresponding Author: A Thakur, Department of Applied Sciences, National Institute of Technical Teachers Training and Research, Bhopal, INDIA.
Received: December 08, 2016; Published: April 05, 2017
Abstract
Present study aim to identify the role of quantum chemical parameters in modeling of HIV-1 inhibition activity of Piperidine-4-Carboxamide CCR5 antagonists. For the purpose a set of 21 Piperidine-4-Carboxamide has been chosen.
Study explores the role of various, quantum parameters like HOMO, LUMO, electron density, Net charge etc. in the anti HIV-1 activity of Piperidine-4-Carboxamide CCR5 antagonist derivatives. Huckel molecular orbital theory is applied to calculate the quantum chemical parameters and multiple regression method is adopted to identify the role of various quantum chemical parameters in modeling the logIC50 activity. Statistics generated from the study shows that none of the parameter having statistical significant value of r in uni-parametric correlation but bi-parametric to tetra-parametric combinations produced the significant value of regression as well as information
Keywords: Quantum chemical descriptors; CCR5 antagonist; Molecular modeling; HIV-1 inhibition
Abbreviations: QSAR: Quantitative Structure Activity Relationship; HIV-1: Human Immunodeficiency Virus 1; EDC: Electron density
on Carbon; EDN: Electron density on Nitrogen; HOMO: Highest occupied molecular orbital; LUMO: Lowest unoccupied molecular orbital
Introduction
Extracting numerical codes of 3D structures in the form of electron density, net charge on atoms, different forms of energies etc. are the important part of computation. These numeric codes in the form of independent variables are used in regression analysis to predict biological function or activity. Quantum descriptors emphasizes mainly on the electronic influence of the compounds. In this way this class of descriptors encoded entirely different properties of the compounds, therefore studied separately.
Quantum chemical methods can be applied to QSAR (quantitative structure activity relationship) by direct derivation of electronic descriptors from the molecular wave function or the electrostatic field. Quantum chemical descriptors are fundamentally different from experimentally measured quantities, although There is however an inherent error associated with the assumptions used in the calculations.
In most cases the direction, but not the magnitude of the error, is known. When using quantum chemistry based descriptors within series of related compounds, the error is considered to be approximately constant throughout these series.
According to classical chemical theory, all chemical interactions are by nature either electrostatic, polar or orbit (covalent) driven. In quantum chemistry, covalent interactions arise from orbital overlap. The interaction of two orbital depends on their energy eigenvalues. Consequently, energies associated with the highest occupied molecular orbital (HOMO), and lowest unoccupied molecular orbital (LUMO) are often good candidates for 2-dimensional descriptors. For example, HOMO might model the covalent basicity of a hydrogen bond acceptor or the LUMO (lowest unoccupied molecular orbital) might model the covalent acidity of the proton of the H bond donor [1]. Further interpretation is possible because the HOMO (highest occupied molecular orbital) energy is related to the ionization potential and is a measure of the molecule's tendency to be attacked by electrophiles. Correspondingly, the LUMO energy is related to the electron affinity and is a measure of a molecule's tendency to be attacked by nucleophiles [1,2]. Furthermore, according to frontier molecular orbital theory, transition state formation involves the interaction between the frontier orbital of reacting species.
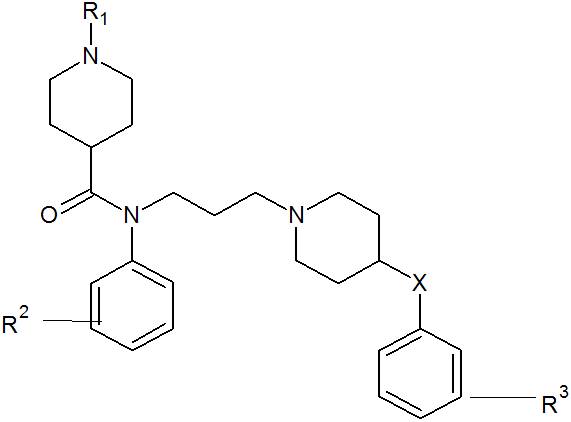
Recent advances of chemokine receptors functioning as HIV-1 have provided a novel strategy for controlling HIV-1 infection [3]. HIV-1 strains that cause the initial infection primarily utilize CC chemokine receptor 5 (CCR5) [4], and CCR5-using (R5) HIV-1 is isolated predominantly during the asymptomatic stage of the infection, which usually persists 5-10 years [5]. CCR5 belongs to the seven-transmembrane G protein-coupled receptor superfamily, and its natural ligands include the CC chemokines macrophage inflammatory protein (MIP)-1R, and MIP-1â], which have been reported to inhibit R5 HIV-1 infection in vitro [6]. the studied CCR5 antagonist derivatives are presented in Table 1 and parent structure of the derivative is presented in figure 1.
| Comp. No. | R1 | R2 | X | R3 |
| 1 | Ms | 3,4-diCl | CH2 | 4-Ms |
| 10 | Ms | 3,4-diCl | NHCO | 4-F |
| 11 | Ms | 3,4-diCl | CH2 | 4-CN |
| 12 | Ms | 3,4-diCl | CH2 | 4-CO2Me |
| 13 | Ms | 3,4-diCl | CH2 | 4-CO2H |
| 14 | Ms | 3,4-diCl | CH2 | 4-CONH2 |
| 15 | Ms | 3,4-diCl | CH2 | 3-CONH2 |
| 16 | Ms | 3,4-diCl | CH2 | 2-CONH2 |
| 17 | Ms | 3,4-diCl | CH2 | 4-CONHMe |
| 18 | Ms | 3,4-diCl | CH2 | 4-CONHt-Bu |
| 19 | Ms | 3,4-diCl | CH2 | 4-CONMe2 |
| 2 | Ms | 3,4-diCl | CH2 | 4-F |
| 20 | Ac | 3,4-diCl | CH2 | 4-CONH2 |
| 21 | Ac | 3-Cl,4-Me | CH2 | 4-CONH2 |
| 3 | Ac | 3,4-diCl | CH2 | 4-F |
| 4 | Ac | H | CH2 | H |
| 5 | Ac | 3,4-diCl | S | 4-F |
| 6 | Ac | 3,4-diCl | SO | 4-F |
| 7 | Ac | 3,4-diCl | SO2 | 4-F |
| 8 | Ac | 3,4-diCl | NH | 4-F |
| 9 | Ms | 3,4-diCl | NHSO2 | 4-F |
Table 1: Various substituent of piperidine-4-carboxamide7 investigated in the present study.
The Quantum descriptors and indicator parameters tested in present study are given in Table 2.
| Comp. No. | HOMO | EDC4 | EDN | IMS | IX |
| 1. | -0.218 | 3.79 | 4.89 | 1 | 1 |
| 10. | -0.216 | 3.80 | 4.89 | 1 | 0 |
| 11. | -0.122 | 3.45 | 5.28 | 1 | 1 |
| 12. | -0.219 | 3.80 | 4.89 | 1 | 1 |
| 13. | -0.220 | 3.79 | 4.89 | 1 | 1 |
| 14. | -0.220 | 3.79 | 4.89 | 1 | 1 |
| 15. | -0.219 | 3.80 | 4.90 | 1 | 1 |
| 16. | -0.219 | 3.79 | 4.89 | 1 | 1 |
| 17. | -0.219 | 3.79 | 4.89 | 1 | 1 |
| 18. | -0.219 | 3.80 | 4.90 | 1 | 1 |
| 19. | -0.220 | 3.79 | 4.89 | 1 | 1 |
| 2. | -0.219 | 3.79 | 4.89 | 0 | 1 |
| 20. | -0.218 | 3.79 | 4.97 | 0 | 1 |
| 21. | -0.213 | 3.88 | 4.97 | 0 | 1 |
| 3. | -0.218 | 3.79 | 4.98 | 0 | 1 |
| 4. | -0.218 | 4.02 | 4.98 | 0 | 1 |
| 5. | -0.216 | 3.79 | 4.98 | 0 | 0 |
| 6. | -0.218 | 3.79 | 4.97 | 0 | 0 |
| 7. | -0.218 | 3.79 | 4.97 | 0 | 0 |
| 8. | -0.183 | 3.79 | 4.97 | 0 | 0 |
| 9. | -0.219 | 3.79 | 4.95 | 0 | 0 |
*HOMO= Highest Occupied Molecular Orbital
EDC4= Electron Density at 4th Carbon
EDCN= Electron Density on Nitrogen
IMS = Indicator parameter for Mesityl group at R1 position in the parent moiety
Ix = Indicator parameter for –CH2- group at X position
Table 2: Quantum chemical parameters and indicator parameters tested in present study.
EDC4= Electron Density at 4th Carbon
EDCN= Electron Density on Nitrogen
IMS = Indicator parameter for Mesityl group at R1 position in the parent moiety
Ix = Indicator parameter for –CH2- group at X position
Table 2: Quantum chemical parameters and indicator parameters tested in present study.
Materials and Methods
There are number of modeling techniques with which QSAR models can be built. Based on the nature of the method used, QSAR models are classified as linear or nonlinear. However, the modeling process does not simply consist of passing data through an algorithm. We cannot directly calculate quantum chemical properties or biological activities it requires to take an indirect route.
As a result, QSAR modeling is a stepwise process consisting of six main steps:
- Selecting Biological activity
- Structure entry and optimization
- Descriptor calculations
- Selection of descriptors
- Model development
- Prediction
Another important step in the QSAR model development process is the consideration of the validity of models. In present study it is totally based on the predictive ability of the model. If a QSAR model is to be used as a guide to possible modifications of molecules to improve their activities, the interpretability of the model assumes a major role.
The 3-D optimized structures obtained from the given steps are used to calculate Quantum chemical descriptors by Applying Huckel Molecular Orbital Theory in ChemSW module of molecular modeling pro software [8].
In proposed study methodology will be adopted is based on aspect of Quantitative Structure Activity Relationship i.e., to develop mathematical model based on relation:
Φ = f(C)
Where,
Φ = Biological activity
C = Structural descriptor/ physicochemical properties
C used in present work are topological parameters, physicochemical properties and other molecular features.
F = Function of structure
Φ = f(C)
Where,
Φ = Biological activity
C = Structural descriptor/ physicochemical properties
C used in present work are topological parameters, physicochemical properties and other molecular features.
F = Function of structure
In the present study, quantum chemical descriptors tested are-
- HOMO (Highest occupied Molecular Orbital)
- LUMO (Lowest Unoccupied Molecular Orbital)
- Net charge of specific atom
- Electron density of specific atom
Along with these quantum descriptors two indicator parameters viz IMS = Indicator parameter for Mesityl group at R1 position in the parent moiety and Ix = Indicator parameter for –CH2- group at X position are also tested.
In the further step quantum chemical descriptors are correlated with the biological activity, to perform this Multiple linear regression (MLR) method is used [9].
Result and Discussion
The QSAR analysis using stepwise multiple linear regression method is performed with Quantum chemical descriptors, Here quantum chemical descriptors are used as independent variable to predict the biological activity.
From the persual of univariate correlation, there are two descriptors showing approximetely same correlation with biological activity. Therefore while testing bivariate combination, both the descriptors are examine with the second descriptor, in other words, in first step of screening two descriptrs are screened. These are Ims and EDN (Electron density at Nitrogen of R1) with correlation value 0.34727 and -0.36306 respectively.
Amongst the various combinations tested with the quantum descriptors only best ones are mentioned in the form of correlation models. Starting from bivariate combination, upto tetravariate combination were tested, no pentavariate combinations are tried because of small dataset of 21 compounds.
As it was mentioned in the description that Ims and EDN (electron density at Nitrogen of R1) both are comparable descriptors in a univariate correlation, with r values 0.34727 and -0.36302 respectively, therefore each quantum descriptor has been tested with Ims as well as with EDN in bivariate combination. But, it is clearly observed from the bivariate combinations, that Ims perform better than EDN in bivariate combination. Therefore, the bivariate combination selected to process for trivariate combination must contain Ims rather than EDN.
Combination with the maximum suitable statistical parameters obtained from the combination of HOMO and EDC4 (electron density at 4th carbon). The successive mathematical models obtained from the step wise regression analysis are given below in the form of Eq (1) to Eq (5) with their statistical parameters.
There are two univariate model found with approximate results, therefore given in the form of Eq (1) and (2)
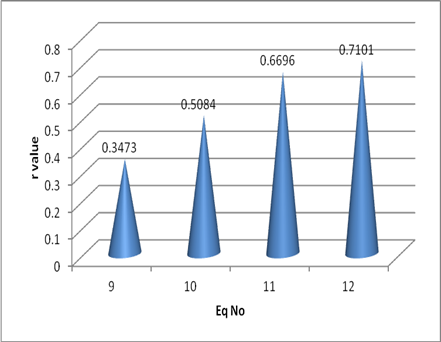
CCR5logIC50 = 0.4297 (±0.2662) Ims + 0.6086 Eq (1)
N = 21, r = 0.3473 Se = 0.6092 F = 2.606
CCR5logIC50 = 0.4297 (±0.2662) Ims + 0.6086 Eq (1)
N = 21, r = 0.3473 Se = 0.6092 F = 2.606
CCR5logIC50 = -2.6514 (±1.5613) EDN + 0.5053 Eq (2)
N = 21, r = -0.363 Se = 0.6053 F = 2.884
N = 21, r = -0.363 Se = 0.6053 F = 2.884
CCR5logIC50 = 0.6627 (±0.2816) Ims - 0.5696 (±0.3113) Ix + 0.8934 Eq (3)
N = 21, r= 0.5084, Se = 0.5747 F = 3.138
N = 21, r= 0.5084, Se = 0.5747 F = 3.138
CCR5logIC50 = 0.895 (±0.268) Ims – 0.683(±0.28) Ix + 3.14 (±1.3)EDC4 – 11.0608 Eq (4)
N = 21 r = 0.6696, Se 0.5101, F = 4.606
N = 21 r = 0.6696, Se 0.5101, F = 4.606
CCR5logIC50 = 11.34 (±8.45) HOMO + 1.004(±0.274) Ims – 0.7305(±0.276)Ix + 5.403(±2.11)EDC4 – 17.25 Eq (5)
N = 21 r = 0.7101, Se 0.4985, F = 4.068
N = 21 r = 0.7101, Se 0.4985, F = 4.068
The relative increase in the r value can be easily observed with the help of the graph shown in the Figure 2.
It is clearly observed from the model obtained from quantum descriptors that EDN produces no significant role in the following steps of regression analysis, therefore it is screened out from the regression analysis, and finally does not appear in the final mathematical model.
The best correlation obtained using Quantum descriptors is 0.7101. This is a significant result, and the model with such result may consider as efficient model for the prediction purpose, and it is valuable for the screening of HOMO and EDC4 from the other Quantum descriptors. These two descriptors are not seems to be important descriptor initially in correlation matrix, but both the descriptors are came out as operational descriptors.
By the persual of Eq (5) Ims, Ix, HOMO and EDC4 are selected as predictive descriptors among the quantum chemical descriptors.
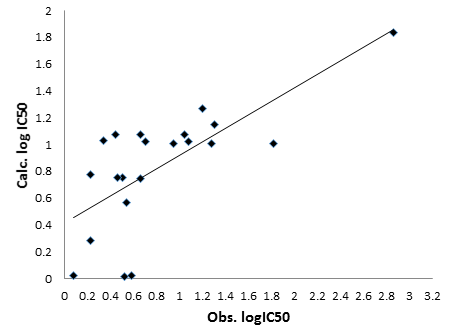
The prediction of Biological activity, i.e., inhibition of 125I-labeled RANTES binding to Chinese hamster ovary (CHO) cells expressing human CCR5 on their surface is calculated or predicted using mathematical model obtained from quantum descriptors and present in Table 3.
| Comp. No. | Obs. logIC50 | Calc. logIC50 | Residual |
| 1. | 0.342 | 1.03 | -0.6876 |
| 10. | 2.863 | 1.837 | 1.0262 |
| 11. | 0.23 | 0.281 | -0.0508 |
| 12. | 0.663 | 1.072 | -0.4093 |
| 13. | 1.82 | 1.007 | 0.8131 |
| 14. | 0.949 | 1.007 | -0.0579 |
| 15. | 0.447 | 1.072 | -0.6253 |
| 16. | 1.079 | 1.018 | 0.0608 |
| 17. | 0.708 | 1.018 | -0.3102 |
| 18. | 1.041 | 1.072 | -0.0313 |
| 19. | 1.279 | 1.007 | 0.2721 |
| 2. | 0.519 | 0.014 | 0.5053 |
| 20. | 0.58 | 0.025 | 0.5549 |
| 21. | 0.544 | 0.568 | -0.0241 |
| 3. | 0.079 | 0.025 | 0.0539 |
| 4. | 1.204 | 1.268 | -0.0639 |
| 5. | 0.23 | 0.778 | -0.5483 |
| 6. | 0.505 | 0.756 | -0.2506 |
| 7. | 0.462 | 0.756 | -0.2936 |
| 8. | 1.301 | 1.152 | 0.1486 |
| 9. | 0.662 | 0.744 | -0.0823 |
Table 3: Experimental and calculated biological activities using Eq (5) for
Piperidine-4-carboxamide derivatives tested in the present study.
The correlation between experimental and calculated activity is present in Figure 3.
The purpose of carrying out this study at last is to obtain highly predictive model, and to test the quantum descriptors and both the objectives are successfully attain. This can be clearly observed from the results mentioned.
Descriptor’s Contribution: The negative coefficient of Ix indicate that the presence of –CH2- group decreases the value of biological activity in a quantitative manner, i.e., Presence of –CH2- is favorable. On the other hand, positive coefficient of Ims leads to increase in the value of biological activity in a quantitative manner i.e., presence of Mesityl group is not favorable.
The positive coefficient of HOMO shows that the value of CCR5logIC50 increases with the increase in the value of HOMO (energy of highest occupied molecular orbital), higher value of CCR5logIC50 is unfavorable. Therefore, a molecule should have such structure which possesses lower value of HOMO. Similarly positive coefficient of EDC4 shows that the value of CCR5logIC50 increases with the increase in the value of EDC4 (Electron density at 4th Carbon), higher value of CCR5logIC50 is unfavorable.
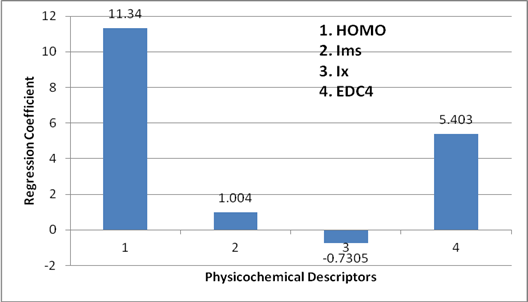
The preference of one descriptor over the other is represented in the form of bar graph (Figure 4). Height of bar is the measure of the dominance of descriptor over the other. It is clearly shown in the (Figure 4) that the HOMO is playing a dominating role over the other descriptors. The second dominant descriptor is EDC4 and the third is Ims and the lowest participation is Shown by the Ix. That means the order of preference among the Quantum descriptor is
HOMO > EDC4 > Ims > Ix
HOMO > EDC4 > Ims > Ix
Conclusion
An exhaustive search performed for the best one-, two-, three-, and four-parameter regression models. As seen from the statistical plots presented, the optimum number of parameters for the correlation equation is four.
Total five QSAR models are developed with different quantum descriptors to assess the predictive power of QSAR models for inhibition activity. It is interesting to note that in the cases, tetra-parametric model is adequate. The study reveals that quantum chemical descriptors are the most important class of descriptors used. In the quantum class of descriptors, energy of highest occupied molecular orbital (HOMO), Electronic density at C4 (EDC4) descriptors, with indicator parameters Ims and Ix give the most significant QSAR model.
The QSAR model in the form of Eq. (5) is contains HOMO and EDC4 with the positive coefficient; it means both the parameters are directly proportional to the value of CCR5logIC50. That means higher value of HOMO and EDC4 raise the value of CCR5logIC50, which implies that the higher concentration is needed for 50% inhibition activity. Therefore, substitution which decreases the energy of HOMO and Electron density at C4 is useful for the compound to make biologically more active.
References
- Sklenar H and Jager J. “Molecular structure–biological activity relationships on the basis of quantum-chemical calculations”. International journal of quantum chemistry16.3 (1979): 467-484.
- Tuppurainen K., et al. “QSAR in Toxicology”. Mutation Research 247.1 (1991): 97-102.
- Berger E A., et al. “Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease”. Annual Review of Immunology 17 (1999): 657-700.
- Fauci AS. “Host factors and the pathogenesis of HIV-induced disease”. Nature 384.6609(1996): 529-534.
- Connor RI., et al. “Change in coreceptor use correlates with disease progression in HIV-1--infected individuals”. The Journal of Experimental Medicine 185.4 (1997): 621-628.
- Cocchi F., et al. “Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells”. Science 270 (1995): 1811-1815.
- Nelson KM., et al. “The Essential Medicinal Chemistry of Curcumin”. Journal of Medicinal Chemistry 60.5 (2017): 1620-1637.
- Molecular modeling Pro: www.labwrench.com.
- Chaterjee S., et al. Regression Analysis by Example 3rd ed. Wiley-VCH: New York (2000).
Citation:
A Thakur., et al. “Role of Quantum Chemical Parameters: Modeling of HIV-1 Inhibition Activity for CCR5 Antagonists”.
Chronicles of Pharmaceutical Science 1.2 (2017): 58-66.
Copyright: © 2017 A Thakur., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.































 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.