Research Article
Volume 2 Issue 2 - 2018
Isolation and Identification of Bacterial Strains in Aerosols Samples from an A Iron Foundry and Study of Their Resistance to Heavy Metals
1Chemical Bioactive Center. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba
2Department of Bioscience Engineering, Environmental Ecology and Microbiology (ENdEMIC), University of Antwerp. Antwerp. Belgium
3Chemistry Department. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba
4Pharmacy's Department. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba
2Department of Bioscience Engineering, Environmental Ecology and Microbiology (ENdEMIC), University of Antwerp. Antwerp. Belgium
3Chemistry Department. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba
4Pharmacy's Department. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba
*Corresponding Author: Ester María Hernández Martínez, Chemical Bioactive Center. Central University "Marta Abreu" of Las Villas. Santa Clara. Villa Clara. Cuba.
Receive: September 06, 2018; Published: September 20, 2018
Abstract
Background: Air pollution poses a significant environmental risk to health. Different investigations have shown the presence of bacteria in the atmosphere. However, few studies of air quality based on microbiological components have been carried out in Cuba.
Objective: Of this research was to isolate and identify bacterial strains present in the workplace atmosphere of an iron factory and determine their resistance to heavy metals.
Methods: Indoor air samples were collected from an iron foundry and viable plate counts were obtained on different selective growth Medias. To identify the bacteria of the pure cultures, their 16S rRNA genes were amplified and sequenced. To study the resistance of the isolated bacteria, different concentrations of heavy metals were tested.
Results: Eleven isolated bacteria belonging to six different species were identified using BLAST program: Pantoea agglomerans, Enterobacter cloacae, Staphylococcus aureus, Bacillus oceanisediminis, Bacillus flexus and Exiguobacterium aurantiacum. All bacterial strains showed an increase in the cell viability at high concentrations of Fe, Zn and Cu and, at low concentrations of Mn compared to the isolates without metal. However, all Pb concentrations tested resulted in an increase of the cell viability for E. aurantiacum, while this was only observed for B. oceanisediminis at high concentrations. S. aureus, B. oceanisediminis, P. agglomerans, B. flexus and E. aurantiacum showed increased cell viability at high concentrations of Sn while P. agglomerans, B. flexus and E. aurantiacum showed this increment at low concentrations.
Conclusion: In the iron smelting industry, the identified bacterial strains showed an increased resistance to the tested heavy metals, indicating that these metals are a driving force in the niche-adaptation of these airborne bacteria and can be harmful to health and even more if it is considered their resistance to tested metals.
Keywords: Bacterial strains; Indoor air; Iron foundry; Heavy metals
Introduction
Environmental pollution is a problem that has increased in recent years, in accordance with the growing trend of population increase and the corresponding increase in urban radius. In addition, the great development of productive and industrial activity has strongly impacted the physical and biological variables of the urban environment [Dales and Vidal 2010; Sanhueza., et al. 2009; USEPA 2015; WHO 2014].
Air pollution poses a significant environmental risk to health, whether in developed or developing countries. According to the latest WHO estimates of the global burden of disease, indoor and outdoor air pollution cause about seven million premature deaths. This is currently one of the largest global health risks, comparable to tobacco-related risks, and only outweighed by the health risks associated with hypertension and nutrition. (WHO 2014)
The main air pollutants are particulate matter (PM), ozone (O3), nitrogen oxides (NOx) (especially nitrogen dioxide and trioxide [NO2 and NO3]) and sulfur oxides (SOx). In the last years, different investigations have observed the presence of bacteria in the atmosphere [Barahona 2010; Després., et al. 2007; Fahlgren., et al. 2010; Smets., et al. 2016]. However, in Cuba few studies on the air quality have yet explored the presence of microorganisms in urban or rural environments. Instead, the investigations that have been carried out, focused on the physical and chemical characterization of the different atmospheric pollutants [Barja., et al. 2011; Núñez., et al. 2014].
Environmental contaminants of biological origin (bioaerosols) are constituted of particles, large molecules, and volatile organic compounds that come from living organisms. In bioaerosols microorganisms and their fragments, toxins and metabolic products can be found [Ghosh., et al. 2015; Fröhlich-Nowoisky., et al. 2016; Maldonado., et al. 2014]. The survival, reproduction and dispersion of biological pollutants in the air depends to a large extent on the conditions of the environment in which they are found. These conditions include the available resources and the key stress factors (such as desiccation, organic and inorganic pollutants and UV irradiation) [Smets., et al. 2016]. Interestingly, not all atmospheric pollutants have a negative effect on the atmospheric bacteria. For instance, nitrogen monoxide (NO), carbon monoxide (CO) andhydrocarbons may have a protective or growth-promoting effect on microorganisms, depending on the species [Barahona 2010; Chandra., et al. 2005; De la Rosa., et al. 2002; Jones and Harrison 2003; Lin and Li 2000]. Yet, dispersal plays also a key role in the abundance of microorganisms in the air and this is dependent on various factors such as turbulent circulation, vehicles, wind, temperature, availability of water, amount of suspended dust, etc.
Heavy metal contamination is a widespread environmental problem because they are non-biodegradable and have the potential to accumulate in human and animal bodies. Most heavy metals are extremely toxic even at low concentrations depending on the solubility of the heavy metal compounds in water [Arora., et al. 2008, Park and Chon 2016]. Also bacteria interaction with metals (Suárez and Reyes 2002). Some heavy metals like copper (Cu), selenium (Se), and zinc (Zn) have been described as essential for maintaining metabolism in living beings. However, for others such as mercury (Hg), lead (Pb) and cadmium (Cd) no biological function has been found [Suárez and Reyes 2002].
In this study, we aimed to explore the impact of heavy metals on bacteria that were isolated from the atmosphere of an iron foundry, to explore whether these bacteria show a resistance to heavy metals.
Materials and Methods
Sampling
Indoor air samples were taken at the iron foundry in Villa Clara (Cuba) using a low volume sampler MCV CPV-8D/A, in which air was bubbled in trypticase soy broth (TSB) in the equipment bubblers, for 8 hours. The samples were collected in sterile plastic vials, transferred to the laboratory, stored at 4°C and then processed.
Indoor air samples were taken at the iron foundry in Villa Clara (Cuba) using a low volume sampler MCV CPV-8D/A, in which air was bubbled in trypticase soy broth (TSB) in the equipment bubblers, for 8 hours. The samples were collected in sterile plastic vials, transferred to the laboratory, stored at 4°C and then processed.
Culturing
A serial dilution of samples was made using Phosphate Buffered Saline (PBS) until a dilution of 10-6 was obtained. 75 μL of 10-4, 10-5, and 10-6 dilutions each were inoculated on trypticase soy agar (TSA; Fluka Analytical, Sigma-Aldrich) and incubated at 37°C for 24h. This temperature was chosen to represent the Cuban climate. Individual colonies of different morphologies were subcultured twice in trypticase soy broth (TSB) at 37°C.
A serial dilution of samples was made using Phosphate Buffered Saline (PBS) until a dilution of 10-6 was obtained. 75 μL of 10-4, 10-5, and 10-6 dilutions each were inoculated on trypticase soy agar (TSA; Fluka Analytical, Sigma-Aldrich) and incubated at 37°C for 24h. This temperature was chosen to represent the Cuban climate. Individual colonies of different morphologies were subcultured twice in trypticase soy broth (TSB) at 37°C.
Identification of strains
To identify the isolates, their 16S rRNA genes were amplified and sequenced. For polymerase chain reaction (PCR) a partial colony was transferred to 10 µL PCR-grade H20 in a PCR tube and microwaved for 2 x 1.5 minutes at 700W. dNTPs, buffer and Taq polymerase (VWR) were added together with universal bacterial primers for the 16S rRNA gene: 27F (5’- AGAGTTTGATCCTGGCTCAG -3’) and 1492R (5’- GGTTACCTTGTTACGACTT -3’) (Eden., et al. 1991). The end concentrations were 1 μM of each primer in a final volume of 25 µL. Negative controls for master mix contamination were included. The amplification program was performed with an initial denaturation step of 2 min at 95°C; followed by 30 cycles of 30 seconds at 95°C, 30 seconds at 55°C and 1 minutes 30 seconds at 72°C; and a final extension step at 72°C for 5 minutes. The PCR products were visualized on a 1% agarose gel and those that showed clear bands were sent for Sanger sequencing with both the 27F and 1492R primer (VIB Genetic Service Facility, Antwerp, Belgium). The resulting sequences were compared to the NCBI database using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/) to classify the selected isolates at species or genus level.
To identify the isolates, their 16S rRNA genes were amplified and sequenced. For polymerase chain reaction (PCR) a partial colony was transferred to 10 µL PCR-grade H20 in a PCR tube and microwaved for 2 x 1.5 minutes at 700W. dNTPs, buffer and Taq polymerase (VWR) were added together with universal bacterial primers for the 16S rRNA gene: 27F (5’- AGAGTTTGATCCTGGCTCAG -3’) and 1492R (5’- GGTTACCTTGTTACGACTT -3’) (Eden., et al. 1991). The end concentrations were 1 μM of each primer in a final volume of 25 µL. Negative controls for master mix contamination were included. The amplification program was performed with an initial denaturation step of 2 min at 95°C; followed by 30 cycles of 30 seconds at 95°C, 30 seconds at 55°C and 1 minutes 30 seconds at 72°C; and a final extension step at 72°C for 5 minutes. The PCR products were visualized on a 1% agarose gel and those that showed clear bands were sent for Sanger sequencing with both the 27F and 1492R primer (VIB Genetic Service Facility, Antwerp, Belgium). The resulting sequences were compared to the NCBI database using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/) to classify the selected isolates at species or genus level.
Heavy metal resistance
The sensitivity of the bacteria to heavy metal ions was determined by growing the isolates in TSB with the added heavy metal. Six different concentrations of iron (Fe), Zn, Cu, Pb, tin (Sn), aluminium (Al) and manganese (Mn) were tested in the form of their salts, iron(II) sulphate heptahydrate (FeSO4∙7H2O), zinc sulphate heptahydrate (ZnSO4∙7H2O), copper(II) sulphate pentahydrate (CuSO4∙5H2O), lead(II) chloride (PbCl2), tin(II) chloride nonahydrate (SnCl2∙9H2O), aluminium sulphate octadecahydrate (Al2(SO4)3∙18H2O) and manganese(II) sulphate monohydrate (MnSO4∙H2O) (see Table 1), taking into account the values of each metal obtained in a 12-hour sampling period [Alejo 2017] (Figure 1).
The sensitivity of the bacteria to heavy metal ions was determined by growing the isolates in TSB with the added heavy metal. Six different concentrations of iron (Fe), Zn, Cu, Pb, tin (Sn), aluminium (Al) and manganese (Mn) were tested in the form of their salts, iron(II) sulphate heptahydrate (FeSO4∙7H2O), zinc sulphate heptahydrate (ZnSO4∙7H2O), copper(II) sulphate pentahydrate (CuSO4∙5H2O), lead(II) chloride (PbCl2), tin(II) chloride nonahydrate (SnCl2∙9H2O), aluminium sulphate octadecahydrate (Al2(SO4)3∙18H2O) and manganese(II) sulphate monohydrate (MnSO4∙H2O) (see Table 1), taking into account the values of each metal obtained in a 12-hour sampling period [Alejo 2017] (Figure 1).
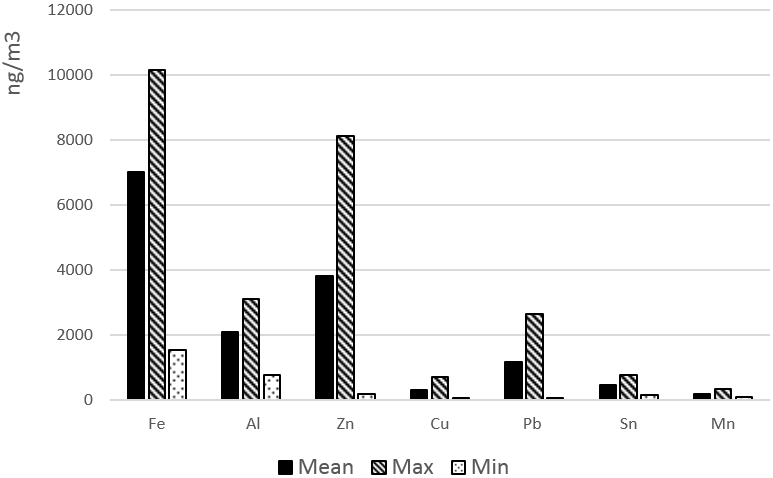
Figure 1: Metal concentrations in iron-foundry air. Mean,
Min and Max values of 12-hour measurements.
| Concentration | FeSO4∙7H2O (mM) | Al2(SO4)3∙18H2O (mM) | ZnSO4∙7H2O (mM) | CuSO4∙5H2O (mM) | PbCl2 (mM) | SnCl2∙9H2O (mM) | MnSO4∙H2O (mM) |
| 1 | 3.6 | 2.3 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| 2 | 2 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 |
| 3 | 0.5 | 0.12 | 0.12 | 0.1 | 0.1 | 0.1 | 0.1 |
| 4 | 0.09 | 0.06 | 0.06 | 0.006 | 0.006 | 0.003 | 0.003 |
| 5 | 0.06 | 0.04 | 0.03 | 0.003 | 0.003 | 0.002 | 0.002 |
| 6 | 0.01 | 0.01 | 0.001 | 0.0006 | 0.0002 | 0.0007 | 0.0009 |
Table 1: Concentrations tested for the heavy metals.
Working solutions of metals were prepared by diluting the stock solutions with TSB 30 minutes prior to use. Each condition was tested in triplicate using a 96-well format with 100 µL final volume per well. In addition, duplicate controls for each metal concentration in absence of bacteria, for each tested isolate in absence of metal, and with pure TSB were included and used for normalization.
After incubation at 37°C for one hour, 10 μL of alamarBlue® (DAL1025, Thermo Fisher Scientific) was added to each well. Absorbance at 570 nm and 600 nm was read after 1h and 4h using the Synergy HTX plate reader (Biotech Instruments, Inc.) and the cell viability was calculated with the following formula:
Results
Three samples were collected from the iron foundry in Villa Clara (Cuba). The samples were grown in trypticase soy agar and 11 isolates were obtained. The identified bacterial strains and their GenBank accession numbers are shown in Table 2.
| Bacterial strain | Name | % Identity | NCBI Accession |
| F1 | Pantoea agglomerans | 99% | KU935452.1 |
| F2 | Enterobacter cloacae | 99% | CP009850.1 |
| F3 | Enterobacter cloacae | 99% | CP009850.1 |
| F4 | Enterobacter cloacae | 99% | CP009850.1 |
| F5 | Pantoea agglomerans | 99% | KU935452.1 |
| F6 | Enterobacter cloacae | 99% | CP009850.1 |
| F7 | Staphylococcus aureus | 99% | CP014444.1 |
| F8 | Bacillus oceanisediminis | 99% | CP015506.1 |
| F9 | Pantoea agglomerans | 97% | KU935452.1 |
| F10 | Bacillus flexus | 99% | KU236365.1 |
| F11 | Exiguobacterium aurantiacum | 99% | KU922496.1 |
Table 1: Identification of bacterial strains and Gen Bank accession.
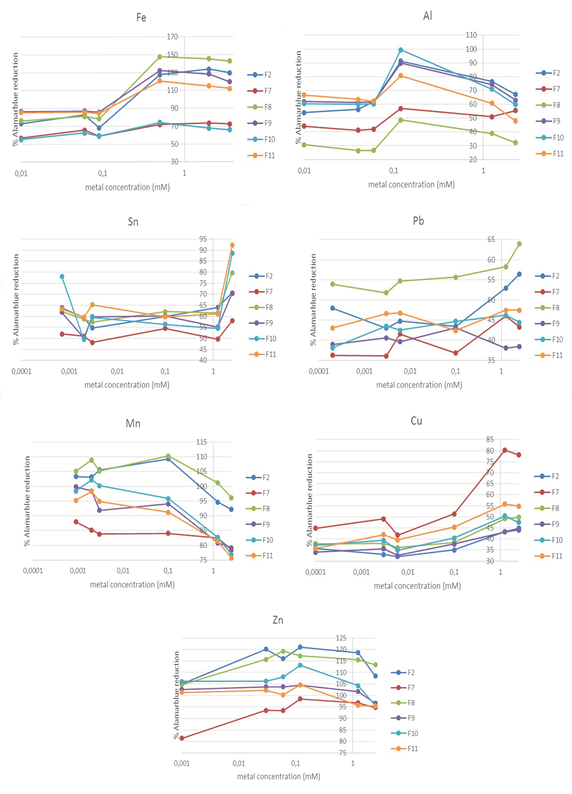
Of the 11 isolates identified, the resistance to heavy metals was determined for F2 (E. cloacae), F7 (S. aureus), F8 (B. oceanisediminis), F9 (P. agglomerans), F10 (B. flexus) and F11 (E. aurantiacum), since the other isolates were similar to F2 and F9.
Iron caused a decrease in viability at the lowest concentrations and an increase of the same at the high concentrations in all tested isolates compared to the isolates without metal (Figure 2) In the case of aluminium, a decrease in viability was observed for all isolates and at all concentrations, with the exception of isolates E. cloacae, B. oceanisediminis and B. flexus, which displayed an increase in viability at a concentration of 0.1 mM. (Figure 2)
E. cloacae showed a decrease in viability in the presence of 0.1 mM tin but a continuous increase at higher concentrations, without achieving the viability of the isolate without the metal. In contrast, for S. aureus and B. oceanisediminis, the viability at the highest concentration (2.5 mM) of tin was higher than that of the isolates without metal, while for the other isolates the viability decreased. For P. agglomerans, B. flexus and E. aurantiacum the viability increased at all concentrations of tin, showing a greater increase at the higher concentration and in the case of P. agglomerans also a very high viability at the lowest concentration. (Figures 2)
In the case of lead, for the bacterial strains E. cloacae, S. aureus, P. agglomerans and Bacillus flexus the viability decreased for all concentrations, in agreement with lead being toxic. In contrast, in the case of B. oceanisediminis it only increased slightly at the highest concentration, whereas for E. aurantiacum it increased slightly for the concentrations of 0.003, 0.006, 1.25 and 2.5 mM (Figures 2)
With manganese, E. cloacae and S. aureus showed a decrease in their cellular viability at the highest concentrations of 1.25 and 2.5 mM, with a slight increase compared to the isolates without metal at all other concentrations. The viability of B. oceanisediminis only decreased at the highest concentration. In contrast, for P. agglomerans and B. flexus a decrease in viability was observed at all concentrations except for 0.0009 mM and 0.002 mM. Finally, the viability of E. aurantiacum decreased from 0.1 mM onwards (Figure 2). At a copper concentration of 1.25 and 2.5 mM, all the bacterial strains showed increased viability compared to isolates without metal. In contrast, their viability decreased at lower copper concentrations, with the exception of S. aureus, B. flexus and E. aurantiacum, for which the viability was also increased at a concentration of 0.1 mM (Figure 2). Lastly, zinc increased the viability of all tested isolates and at all concentrations. (Figure 2).
Discussion
In an iron smelting industry in the province of Villa Clara bacteria belonging to the phyla Proteobacteria and Firmicutes were identified. The phylum Firmicuteswas most frequently represented with genera such as Bacillus, Staphylococcus and Exiguobacterium. These results agree with those obtained by [Di Giorgio., et al. 1996 and Méndez., et al. 2015], who reported that gram positive bacteria tend to be dominant in the cultivable fraction of air samples. These gram positive genera are more resistant to dry or adverse conditions because they have a thicker and peptidoglycan-rich cell wall [Atlas and Bartha 1997; De la Rosa., et al. 2002]. Also, the most abundant genus was Bacillus, which has the ability to form endospores, a latent structure that ensures its survival under stress conditions such as the environmental factors found in the atmosphere [Nicholson 2000].
The phylum Proteobacteria is a major phylum of gram-negative bacteria. The genera identified from the isolates were Enterobacter and Pantoea, belonging to the group of Gammaproteobacteria. Bacteria of the genus Enterobacter are responsible for a wide variety of diseases such as nosocomial infections in neonates and immunosuppressed patients and are difficult to treat because they have developed resistance to a large number of antibiotics [Correa., et al. 2012; Walterson and Stavrinides 2015]. Pantoea sp. are gram negative bacilli belonging to the family Enterobacteriaceae [Sánchez., et al. 2006]. The term Pantoea derives from the Greek word Pantoios meaning "of all types and sources" describing the wide geographical and ecological distribution of bacteria belonging to this genus [Rostenberge., et al. 2006; Sánchez., et al. 2006].
Bacteria have developed various resistance mechanisms to tolerate the harmful effects of toxic metals [Silver and Phung 2005]. Among them are mainly those that involve: a) cellular components that capture the ions, neutralizing their toxicity, b) enzymes that modify the redox state of the metals or metalloids, turning them into less toxic forms, and c) membrane transporters that expel the harmful species from the cytoplasm [Cervantes., et al. 2006].
Several heavy-metals are essential cofactors required in trace concentrations by bacteria (nanomolar range); however, they are toxic in higher concentrations [Marrero., et al. 2010]. Iron is an essential element for practically all living organisms, which require it for important cellular functions such as DNA synthesis, respiration and detoxification of free radicals [Aguado., et al. 2012].
Recently it has been described that all the Cation Diffusion Facilitator (CDF) proteins that have been studied so far in bacteria are involved in resistance to Zn (II) and other heavy metal cations [Marrero., et al. 2010]. One of the genera identified in our research and that is resistant to the greatest variety of metals is the genus Bacillus. The presence of this genus in contaminated sites is widely reported in the literature [Montenegro 2007]. Members of this genus produce organic acids capable of binding the metals in the soil. Bacteria are capable of generating extracellular polymers that sequester metals and intervene in the processes of mobilization and immobilization of these elements. The secretion of siderophores which specifically trap Fe3+ ions important for the growth of microorganisms has also been extensively studied [Fingerman and Nagabhushanan 2005; Suarez and Reyes 2002].
Several studies have demonstrated the tolerance of Bacillus sp to environments contaminated with metals such as Pb as well as Cu, Al, Mn and Zn [Herrera 2014; Murthy., et al. 2012]. In (1986) Nakajima and Sakaguchi already reported the ability of these bacteria to accumulate metals. Other studies also reported resistance of Bacillus sp. to various heavy metals [Ali., et al. 2009; Cañizares 2000; Dhal., et al. 2013].
Bacteria that are known to adsorb heavy metals include genera of Bacillus [Nagashetti., et al. 2013; Vijayaraghavan and Yun 2008]. Although there were some studies on the removal of heavy metals by Exiguobacterium sp. [Alam and Ahmad 2013; Batool., et al. 2014], Cd and Pb biosorption by these bacteria, especially when isolated from contaminated sites and under diverse environmental conditions, was not investigated. Alam and Ahmad (2013) investigated Cd, Nickel (Ni), Cu, and Zn biosorption from aqueous solutions by Exiguobacterium sp. ZM-2, but biosorption was not tested in mixed metal solutions. The selective biosorption of Cd and Pb by Exiguobacterium sp. was observed by Park and Chon (2016).
According to the literature, Staphylococcus aureus is resistant to metals by the mechanism of expulsion by type P ATPase (CadA) (Ji., et al. 1994; Novick., et al. 1979; Nucifora., et al. 1989; Yoon., et al. 1991) and by the CDF-type (RzcB) expulsion mechanism [Xiong., et al. 1998].
Several studies [Adelaja and Keenan 2012; Campos., et al. 1995; Dash., et al. 2013; Kavamura and Esposito 2010; Sinha., et al. 2012; Wang., et al. 1989] reported resistance of Enterobacter cloacae to heavy metals. These genera have been reported in the literature as resistant to multiple antibiotics [Alósa 2015; Castañeda., et al. 2009; Martínez., et al. 2010; Vanegas., et al. 2009] and as observed here, also show resistance to various heavy metals. The correlation of resistance to heavy metals and antibiotics between clinical and environmental isolates is important and has been well studied. Experimental investigations indicate that increased exposure to these metals alters several parameters of the immune system and lead to increased susceptibility to infections, autoimmune diseases and allergic manifestations as well as damage to the Central Nervous System [Atencio., et al. 2005].
Genes have been reported, not associated with mobile elements, which can encode determinants of resistance to antibiotics and heavy metals. This chromosomal arsenal of resistance elements together with a membrane of low permeability have been proposed as responsible for a multi-resistant intrinsic phenotype that is independent of the environment in which some bacteria live [Cervantes., et al. 2006; Ramakrishna and Kefeng 2011].
Thus, the indiscriminate use of antibiotics, for human consumption, as well as for veterinary and agricultural purposes, is the main factor for the selection of bacteria resistant to these chemical compounds. In addition, the high environmental contamination by heavy metals favors the selection of strains resistant to antibiotics, when the genes responsible for these resistances are on the same plasmid [Paniagua., et al. 2003].
Storage at 4°C may have selected for psychrophilic bacteria, if present, but incubation at 37°C will have promoted the growth of species representative for the hot cuban climate. In addition, more species would have been detected with a sequencing approach, but the cultivable fraction of the iron foundry air microbiome was of higher interest for metal-sensitivity testing.
Conclusion
The genera identified from industrial air samples were Enterobacter sp., Pantoea sp., Staphylococcus sp., Bacillus sp., and Exiguobacterium sp., Bacillus oceanisediminis and Exiguobacterium aurantiacum proved to be the bacteria resistant to the largest variety of heavy metals at almost all concentrations.
In the iron smelting industry, the identified bacterial strains showed an increased resistance to the tested heavy metals, indicating that these metals are a driving force in the niche-adaptation of these airborne bacteria and can be harmful to health and even more if it is considered their resistance to tested metals.
Acknowledgements
This research was funded by the VLIR Program, Project Characterization and analysis of particulate compounds in multiple workplace atmospheres, University of Antwerp, Belgium. The authors thank Prof. Dr. Sarah Lebeer and Serena Moretti, Department of Bioscience Engineering, Environmental Ecology and Microbiology (End EMIC) for their help.
This research was funded by the VLIR Program, Project Characterization and analysis of particulate compounds in multiple workplace atmospheres, University of Antwerp, Belgium. The authors thank Prof. Dr. Sarah Lebeer and Serena Moretti, Department of Bioscience Engineering, Environmental Ecology and Microbiology (End EMIC) for their help.
References
- Adelaja O., et al. “Tolerance of TBT-resistant bacteria isolates to methylmercury”. Research Journal of Environmental Sciences 6.1 (2012): 1-13.
- Alam MZ., et al. “Multi-metal biosorption and bioaccumulation by Exiguobacterium sp. ZM-2”. Annals of Microbiology 63 (2013): 1137-1146.
- Alejo D. “Chemical composition of PM10 collected at workplace atmosphere in an iron foundry”.Trends in Green Chemistry 3 (2009): 2.
- Ali N., et al. “Physicochemical characterization and bioremediation perspective of textile effluent, dyes and metals by indigenous bacteria”. Journal of hazardous materials 164 (2009): 322-328.
- Alósa JI. “Antibiotic resistance: A global crisis”. Enfermedades Infecciosas y Microbiología Clínica 33 (2015): 692-699.
- Arora M., et al. “Heavy metal accumulation in vegetables irrigated with water from different sources”. Food Chemistry 111(2007): 811-815.
- Atlas R., et al. “Microbial Ecology: Fundamentals and Applications. San Francisco, CA: Benjamin/Cummings (1997).
- Aguado GA., et al. “Impact of the microbial siderophores and phytosiderophores on the iron assimilation by plants: a synthesis”. Revista Fitotecnia Mexicana 35.1 (2011): 9-21.
- Barahona S. “Microbiological study of the atmospheric particulate material of Santiago using molecular biology tools.” Master's Thesis. University of Chile, Santiago, Chile (2010).
- Barja B., et al. “Atmospheric particulate matter fractions measured at Camagüey, Cuba”. Preliminary results Óptica Pura y Aplicada 44.1 (2011): 115‐125.
- Batool R., et al. “Comparative study of Cr (VI) removal by Exiguobacterium sp. in free and immobilized forms”. Bioremediation Journal 18 (2014): 317-327.
- Bozo L., et al. “Biomarkers of chemical contamination in microbial communities”. Interciencia 32.1 (2007).
- Campos J., et al. “Hexavalent chromium reduction by a chromate resistant Bacillus strain”. Anton Leeuw 68 (1995): 203-208.
- Cañizares R. “Biosorption of heavy metals through the use of microbial biomass.Saudi Journal of Biological Sciences 42 (2000): 131-143.
- Castañeda Y., et al. “Antibiotic susceptibility of faecal pollution indicator bacteria isolated from marine water and sediments at beaches on Margarita Island, Venezuela”. Saber 21.1 (2009): 12-19.
- Chandra P., et al. “Assessment of microbial (bacteria) concentrations of ambient air at semi-arid urban region: influence of meteorological factors”. Applied Ecology and Environmental Research 3(2005): 139-149.
- Cervantes C., et al. “Microbial interactions with heavy metals”. Revista latinoamericana de microbiología 48.2 (2006): 203-210.
- Correa V., et al. “The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector”. Applied and Environmental Microbiology 78 (2012): 6327-6336.
- Dhal B., et al. “Chemical and microbial remediation of hexavalent chromium form contaminated soil and mining/metallurgical solid waste: A review”. Journal of hazardous materials 250.251 (2013): 272-291.
- Dales R., et al. “Air pollution and hospitalization for venous thromboembolic disease in Chile”. International Society on Thrombosis and Haemostasis 8.4 (2010): 669-674.
- Dash H., et al. “Marine bacteria: potential candidates for enhanced bioremediation”. Applied Microbiology and Biotechnology 97 (2013): 561-571.
- De la Rosa M., et al. “The air: habitat and means of transmission of microorganisms”. Observatorio Medioambiental 5 (2012): 375-402.
- Després V., et al. “Molecular genetics and diversity of primary biogenic aerosol particles in urban, rural, and high-alpine air”. Biogeosciences Discussions 4.1 (2007): 349-384.
- Di Giorgio C., et al. “Atmospheric pollution by airborne microorganisms in the city of Marseilles”. Atmospheric 2310.95 (1996).
- Eden P., et al. “Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA”. International Journal of Systematic and Evolutionary Microbiology 41 (1991): 324-325.
- Fahlgren C., et al. “Annual variations in the diversity, viability, and origin of airborne bacteria”. Applied and Environmental Microbiology 76.9 (2010): 3015-3025.
- Fingerman M., et al. “Bioremediation of aquatic and terrestrial ecosystems”. Science Publishers: USA (2005).
- Fröhlich J., et al. “Bioaerosols in the Earth system: Climate, health, and ecosystem interactions”. Atmospheric Research Volume 182 (2016): 346-376.
- Ghosh B., et al. “Review of bioaerosols in indoor environment with special reference to sampling, analysis and control mechanisms”. Environment International 85(2015): 254-272.
- Ji G., et al. “Arsenate reductase of Staphylococcus aureus plasmid pI258”. Biochemistry 33(1994): 7294-7299.
- Jones A., et al. “The effects of meteorological factors on atmospheric bioaerosol concentrations”. Science of the Total Environment 326.1(2003): 151-180.
- Kavaruma V., et al. “Biotechnological strategies applied to the decontamination of soils polluted with heavy metals”. Biotechnology advances 28(2010): 61-69.
- Lin W., et al “Associations of fungal aerosols, air pollutants, and meteorological factors”. Aerosol Science and Technology 32(2010): 359-368.
- Maldonado M., et al. “Bioaerosols and air quality assessment in two hospitals located in León, Guanajuato, Mexico”. Revista internacional de contaminación ambiental 30.4 (2014): 351-363.
- Marrero J., et al. “Molecular mechanisms of resistance to heavy metals in bacteria and their applications in bioremediation”. Revista CENIC Ciencias Biológicas 41.1 (2010): 67-78.
- Martínez A., et al. “Resistance to antibiotics and heavy metals in bacteria isolated from the Almendares River”. Revista CENIC. Ciencias Biológicas 41(2010): 1-10.
- Méndez CA., et al. “Identification of bacteria and fungi in the air of Neiva, Colombia”. Revista de Salud Pública 17.5 (2015): 728-737.
- Moraga R., et al “Resistance to heavy metals in bacteria isolated from Iquique bay”. Investigaciones marinas 31(2003): 91-95.
- Murthy S., et al. “Biosorption of lead by Bacillus cereus isolated from industrial effluents”. British Biotechnology Journal 2(2012): 73-84.
- Nagashetti V., et al. “Biosorption of heavy metals from soil by Pseudomonas aeruginosa.” International Journal of Innovative Technology and Exploring 2(2012): 22-24.
- Nakajima A., et al. “Selective accumulation of heavy metals by microorganisms”. Applied Microbiology Biotechnology 24(1986): 59-64.
- Nicholson W., et al. “Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments”. Microbiology and Molecular Biology Reviews 64.3 (2000): 548-572.
- Novick RP., et al. “Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps”. Plasmid 2(1979): 109-129.
- Nucifora G., et al. “Cadmium resistance results from a cadmium-efflux ATPase”. Proc Natl Acad Sci USA 86(1989): 3544-3548.
- Núñez V., et al. “Emissions to the atmosphere of particulate material from sugar mills and sugar refineries in the province of Villa Clara”. ICIDCA sobre los derivados de la caña de azúcar 48.2 (2014): 69-74.
- Paniagua GL., et al. “Resistance to antibiotics and heavy metals in clinical strains of Staphylococcus aureus”. Revista Médica del Hospital General de Mexico S.S 66.2 (2003): 13-21.
- Park JH., et al. “Characterization of cadmium biosorption by Exiguobacterium sp. isolated from farmland soil near Cu-Pb-Zn mine”. Environmental Science and Pollution Research 23:11814-11822.
- Ramakrishna W., et al. “Effect of multiple metal resistant bacteria from contaminated lake sediments on metal accumulation and plant growth”. Journal of Hazardous Materials 189 (2011): 531-539.
- Rostenberge V., et al. “The clinical picture of neonatal infection with Pantoea species”. Japanese journal of infectious diseases 59(2006): 120-121.
- Sanchéz M., et a.l “Bacteremia by Pantoea agglomerans. Anales de Medicina Interna 23(2006): 250-251.
- Sanhueza P., et al “Particulate air pollution and health effects for cardiovascular and respiratory causes in Temuco, Chile: a wood-smoke-polluted urban area”. Journal of the Air & Waste Management Association. 59.12 (2009): 1481-1488.
- Silver S., et al. “A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. Microbiol”. Journal of Biotechnology 32(2005): 587-605.
- Sinha A., et al. “Mercury bioremediation by mercury accumulating Enterobacter sp cells and its alginate immobilized application”. Biodegradation 23.1(2012): 25-34.
- Smets W., et al. “Airborne bacteria in the atmosphere: presence, purpose, and potential. Atmos”. Environ 139(2016): 214-221.
- Suárez P., et al. “The incorporation of heavy metals in bacteria and their importance to the environment”. Interciencia Journal 27(2012): 160-164.
- USEPA. “United States Environmental Protection Agency Supplemental Environmental Projects (SEP) Policy 2015 Update” (2015).
- Vanegas M. “Antibiotic resistance of bacteria isolated from biofilms in a food processing plant”. Revista MVZ Córdoba 14.2 (2009): 1677-1683.
- Vijayaraghavan K “Bacterial biosorbents and biosorption”. Biotechnology Advances 26(2015): 266-291.
- Walterson AM., et al. “Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae”. FEMS Microbiology Reviews 39(2015): 968-984.
- Wang P., et al. “Isolation and characterization of an Enterobacter cloacae strains that reduces hexavalent chromium under anaerobic conditions”. Applied and Environmental Microbiology 55.7(1989): 1665-1669.
- World Health Organization. 2014. Children: reducing mortality.
- Xiong A., et al. “Molecular haracterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus.” Journal of Bacteriology 180.16(1989): 4024-4029.
- Yoon KP., et al. “A second gene in the Staphylococcus aureus cadA cadmium resistance determinant of plasmid pI258”. Journal of Bacteriology 173(1991): 7636-42.
Citation:
Ester María Hernández Martínez., et al. “Isolation and Identification of Bacterial Strains in Aerosols Samples from an A Iron
Foundry and Study of Their Resistance to Heavy Metals”. Pulmonary Research and Respiratory Care 2.2 (2018): 158-168.
Copyright: © 2018 Ester María Hernández Martínez., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.





























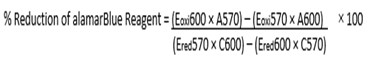
 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.