Research Article
Volume 1 Issue 2 - 2017
Fungi Isolated from Municipal Solid Waste Soil and Rotten Fruit Tested for their Ability to Decolorize Anthraquinonic (Basic Blue) Dye
Microbial Biodiversity Directorate, Ethiopian Biodiversity Institute, Addis Ababa, Ethiopia
*Corresponding Author: Genene Tefera, Microbial Biodiversity Directorate, Ethiopian Biodiversity Institute, Addis Ababa, Ethiopia.
Received: September 05, 2017; Published: September 22, 2017
Abstract
Anthraquinone dyes represent the second largest class of textile dyes, after azo dyes and are used extensively in the textile industry due to their wide array of color shades, ease of application and minimal energy consumption. Anthraquinonic Dye is extensively used in different textile industries and is toxic, mutagenic and carcinogenic in nature. Being recalcitrant, if not treated, they will remain in nature for extended period of time. Large numbers of micro flora including bacteria, fungi, algae and yeasts have been explored to decolorize and degrade dyes. The aim of this study was to screen, identify and evaluate potential fungal species decolorizing ability of Anthraquinonic (Basic Blue) Dye. Twenty five fungal strains isolated from municipal solid waste and rotten fruit using BiOLOG MicroStaion. Among the fungal isolates tested, only 4 fungi strain showed 73-92% transmittance measured in turbidimeter in decolorizing Anthraquinonic dye, these are Candida multisgmis, Pichia guilliermonndi, Rhodotrula aurantica A. Trichosporon begelli B. Four species have 34-42% decolorizing ability, Hypopichia burtoni A, Rhodotrula aurentica B, Wingia robosta, Zygo saccaromycees fermentati, 9 species have 16-29% decolorizing ability, Fusarium udum, Deberomyces hanseni, Pichia mexicana, kluyveromyces delphensis, Pencilium argillaceum,Pencilium rotifotrii, Pichia mexicana,Trichoderma aureoviride, and 9 fungal species do not have any decolorizing effect. Therefore, Candida multisgmis, Pichia guilliermonndi, Rhodotrula aurantica A, were superior in laboratory experiment in decolorization that would be a good ecofriendly candidate in textile colored water effluent treatment after small scale evaluation.
Keywords: Anthraquinonic; Dye; Decolorization; Fungi; Textile
Introduction
Rapid industrialization has necessitated the manufacture and use of different chemicals in day-to-day life (Moorthi 2007, Baljeet 2011). The textile industry extensively uses synthetic chemicals as dyes. Over 0.7 million tons of synthetic dyes are produced annually, worldwide About 100,000 of the chemicals in use as component of synthetic dyes (Rafi 1990). More than 3000 different varieties of azo dyes are used in the textile, paper, food, cosmetics and pharmaceutical industries (Maximo., et al. 2003). In Ethiopia, out of the sixteen textile processing units, thirteen factories are run by the private sector, two are operated by the state and one is managed by a joint venture between the government and a Turkish investor. Dyes and chemicals form the most important part of the supply side of this textile finishing industry. Ethiopian textile processing units consumed about 14,250,406 kg of various types of dyes and chemicals in 2011 with their current production capacity. Anthraquinone dyes represent the second largest class of textile dyes, after azo dyes and are used extensively in the textile industry due to their wide array of color shades, ease of application and minimal energy consumption (Aspland, 1997).
Anthraquinone dyes are resistant to degradation due to their fused aromatic structure, which remain colored for a long time. Most of these dyes are toxic, carcinogenic and mutagenic. Decolorization of anthraquinone dye has received much attention due to recalcitrant nature. Active dyes are not readily removed by typical wastewater treatment processes due to their inherent properties, such as stability and resistance towards light or oxidizing agents (Siddiqui., et al. 2010). The excessive discharge of the effluents from the textile industries contains toxic chemicals such as anthraquinone dye adversely affect the natural resources, soil fertility, and aquatic organisms and disturb the integrity of the ecosystem (Mester and Tien 2000; Puvaneswari., et al. 2006) by alters the pH, increases the biochemical oxygen demand (BOD) and chemical oxygen demand (COD), and greatly affect water quality. Discharge of highly colored dye effluents is aesthetically displeasing and cause considerable damage to the aquatic life. Moreover, it is very difficult to treat textile industry effluents because of their high BOD, chemical oxygen demand (COD), heat, color, pH and the presence of metal ions. Among various industries, the textile dying industries discharge large volume of waste water after dyeing process (Zolinger., et al. 1987).
The textile finishing generates a large amount of wastewater containing dyes and represents one of the largest causes of water pollution (Bhatti., et al. 2008), as 10–15% of dyes are lost in the effluent during the dyeing process (Zollinger, 1991). The traditional textile finishing industry consumes about 100 L of water to process about 1 kg of textile material. Due to increasingly stringent environmental legislation, the textile industry is seeking to develop effective waste water remediation technologies, especially those that allow color removal that is largely unaffected by conventional treatment systems (O’Neill., et al. 1999). Although several physical and chemical treatment methods such as precipitation, coagulation, adsorption, flocculation, flotation, electrochemical destruction, reverse osmosis (RO) process and mineralization and decolonization process (Gogate and Pandit, 2004) have some disadvantages such as cost, time, and release of residues, produce large amounts of sludge. All these techniques are minimizing the toxicity level not to neutralize the toxicity (Cooper, 1993). Also Dye removal from wastewater with traditional physiochemical methods usually require the addition of environmental hazardous chemical additives (Robinson.,. et al. 2001).
Considering drawbacks in afore-mentioned treatments, microbial remediation techniques have gained much attention in the last few decades. Microbial decolorization and degradation is an environment friendly and cost-competitive substitute to different conventional treatment technologies (Ansy., et al. 2002; Verma and Madamwar, 2003; Gogate and Pandit, 2004). Microorganisms reduce dyes by bio sorption (Fu and Viraraghavan, 2000), biodegradation (Conneely., et al. 1999) and enzymatic mineralization (Lignin peroxidase, Manganese peroxidase, Manganese independent peroxidase and laccases (Wesenberg., et al. 2003; Pointing and Vrijmoed, 2000). Immobilization of living microorganisms has been described as valuable in biological wastewater treatment (Katzbauer., et al. 1995; Siddiqui., et al. 2009). Application of immobilized living biomass of fungal strains have been proved more practical than the cell-free system, specifically when they expected to show adsorption as well as enzymatic capabilities of dyes degradation (Lin., et al. 2003; Rojek., et al. 2004). Sludge treatment of wastes is often an effective and highly economic system for reducing organic pollutants in wastewater (Siddiqui., et al. 2010). The reduced forms of dyes are further mineralized into simpler energy source. Some microorganisms including bacteria, fungi and algae, can degrade or absorb a wide range of dyes (Robinson., et al. 2001).The biological mode of treatment of dye bath effluents offers distinct advantages over the conventional modes of treatment. Therefore, the main objective of this work is selecting potential ecofriendly fungi species that are able to decolorize Anthraquinonic (Basic Blue) textile dye.
Material and Methods
Study area and Sample Collection
Two hundred and sixty soil samples that have been exposed to municipal solid waste contamination for a long time were collected from Bahirdar, Gonder, Jimma, Mekele, Diredewa, Adama, Awaday through drillings at 9 different points distributed at the center and periphery of municipal solid waste landfill area or damping site, at 0- 5-and 10-cm depths from 20m2 of 24 quadrants (4m x 5m) of major city. Approximately 5g of soil was taken from each depth of sampling point a total of 45g composite soil per sample were stored in sterile sample tube and hundred pieces of rotten fruit waste were collected from fruit growing area around upper and lower Awash and Benishangule Gumuz, and transported using sterile sample tubes and ice-box to Addis Ababa Microbial Biodiversity Directorate laboratory in Ethiopian Biodiversity Institute and kept in +4oC, until processed.
Two hundred and sixty soil samples that have been exposed to municipal solid waste contamination for a long time were collected from Bahirdar, Gonder, Jimma, Mekele, Diredewa, Adama, Awaday through drillings at 9 different points distributed at the center and periphery of municipal solid waste landfill area or damping site, at 0- 5-and 10-cm depths from 20m2 of 24 quadrants (4m x 5m) of major city. Approximately 5g of soil was taken from each depth of sampling point a total of 45g composite soil per sample were stored in sterile sample tube and hundred pieces of rotten fruit waste were collected from fruit growing area around upper and lower Awash and Benishangule Gumuz, and transported using sterile sample tubes and ice-box to Addis Ababa Microbial Biodiversity Directorate laboratory in Ethiopian Biodiversity Institute and kept in +4oC, until processed.
Screening and Isolation of fungi, associated Municipal soil waste and rotten fruit
The samples’ contents were shaken and serially diluted up to 10-6 and cultured through spread plate techniques on malt extract agar, Czapek’s dox agar, Rose Bengal agar and potato dextrose yeast agar. Rotten mango was mashed with a vortex mixer in the test tube and inoculant transferred into potato dextrose yeast agar media by swab. All the plates were incubated at 28°C for 24-96h in digital incubator. The mixed fungal growth was isolated and sub-cultured until pure colonies were obtained for identification and characterization by BiOLOG Micro Station.
The samples’ contents were shaken and serially diluted up to 10-6 and cultured through spread plate techniques on malt extract agar, Czapek’s dox agar, Rose Bengal agar and potato dextrose yeast agar. Rotten mango was mashed with a vortex mixer in the test tube and inoculant transferred into potato dextrose yeast agar media by swab. All the plates were incubated at 28°C for 24-96h in digital incubator. The mixed fungal growth was isolated and sub-cultured until pure colonies were obtained for identification and characterization by BiOLOG Micro Station.
Identification of yeast from municipal solid and rotten fruit using BiOLOG Micro Station
Pure yeast isolates grown on potato dextrose yeast agar were transferred to BiOLOG Universal Growth agar (BUY) and incubated at 26°C for 48h. Pure colony of yeast suspension were prepared in 9 mL sterile distilled water and adjusted to 47+2T using BiOLOG Turbidimeter. 100 µL of inoculum was dispensed using digital pipetter to each of 96 wells of yeast Micro Plate (YT) tagged with different carbon source and incubated at 26°C 24-72h. The YT microplate measures both metabolic reactions as well as turbidity growth to produce identifications. YT Micro Plate is configured with both oxidation tests and assimilation tests. The first 3 rows of the panel (rows A-C) contain carbon source oxidation tests using tetrazolium violet as a colorimetric indicator of oxidation. The next five rows of the panel (rows D-H) contain carbon source assimilation tests. Results from these tests were scored turbidimetrically.
Pure yeast isolates grown on potato dextrose yeast agar were transferred to BiOLOG Universal Growth agar (BUY) and incubated at 26°C for 48h. Pure colony of yeast suspension were prepared in 9 mL sterile distilled water and adjusted to 47+2T using BiOLOG Turbidimeter. 100 µL of inoculum was dispensed using digital pipetter to each of 96 wells of yeast Micro Plate (YT) tagged with different carbon source and incubated at 26°C 24-72h. The YT microplate measures both metabolic reactions as well as turbidity growth to produce identifications. YT Micro Plate is configured with both oxidation tests and assimilation tests. The first 3 rows of the panel (rows A-C) contain carbon source oxidation tests using tetrazolium violet as a colorimetric indicator of oxidation. The next five rows of the panel (rows D-H) contain carbon source assimilation tests. Results from these tests were scored turbidimetrically.
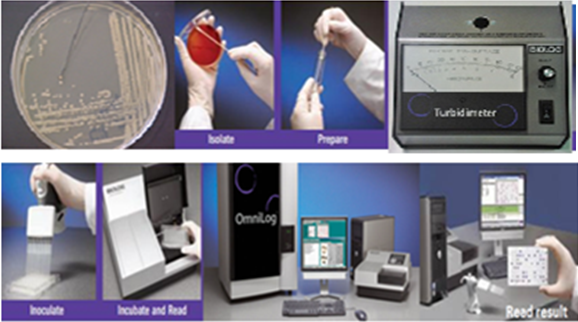
The last row of the panel (row H) has wells that contain 2 carbon sources. These wells test for the co-utilization of various carbon sources with D-xylose.YT MicroPlate was read by the Micro Station reader at 24h, 48h, and 72h at a single wavelength of 590 nm. The BiOLOG software MicroLog3 ver. 4.20.05 compared the results obtained with the test strain to the database and provided identification based on distance value of match and separation score produces similarity index value and probability for species identification (BiOLOG 1993) [Figure.1].
Identification of filamentous fungi from Municipal solid waste soil using BiOLOG Micro Station
Filamentous fungi were isolated and screened on malt extract agar, Czapek’s dox agar, Rose Bengal agar, potato dextrose yeast agar were stained by lactophenol cotton blue in order to confirm to which genera the fungi is belonged to then pure filamentous fungi were transferred into BUY agar media and incubated at 26°C for 48-96h. Pure filamentous fungi suspension were prepared using 15 mL filamentous fungi inoculum fluid (FF IF) and adjusted to 75 + 2T using BiOLOG Turbidimeter.100 µ-L of inoculum was dispensed using digital pipetter to each of 96 wells of filamentous fungi MicroPlate (FF) tagged with different carbon source and incubated at 26°C 24-240h. After incubation, the FF micro plate measured both metabolic reactions as well as turbidity growth to produce identifications. Filamentous fungi MicroPlate (FF) was read by the Micro Station reader at 24h, 48h, and 72-240h at a single wavelength of 590 nm. The BiOLOG software MicroLog3 ver. 4.20.05 compared the results obtained with the test strain to the database and provided identification based on distance value of match and separation score produces similarity index value and probability for species identification (BiOLOG 1993) [Figure 1].
Filamentous fungi were isolated and screened on malt extract agar, Czapek’s dox agar, Rose Bengal agar, potato dextrose yeast agar were stained by lactophenol cotton blue in order to confirm to which genera the fungi is belonged to then pure filamentous fungi were transferred into BUY agar media and incubated at 26°C for 48-96h. Pure filamentous fungi suspension were prepared using 15 mL filamentous fungi inoculum fluid (FF IF) and adjusted to 75 + 2T using BiOLOG Turbidimeter.100 µ-L of inoculum was dispensed using digital pipetter to each of 96 wells of filamentous fungi MicroPlate (FF) tagged with different carbon source and incubated at 26°C 24-240h. After incubation, the FF micro plate measured both metabolic reactions as well as turbidity growth to produce identifications. Filamentous fungi MicroPlate (FF) was read by the Micro Station reader at 24h, 48h, and 72-240h at a single wavelength of 590 nm. The BiOLOG software MicroLog3 ver. 4.20.05 compared the results obtained with the test strain to the database and provided identification based on distance value of match and separation score produces similarity index value and probability for species identification (BiOLOG 1993) [Figure 1].
Anthraquinonic (Basic Blue) dye degradation and decoloring test using fungi
Anthraquinonic (Basic Blue) dye immersed in 10 mL sterile distilled water until it is adjusted to 0% Transmittance using BiOLOG turbidimeter and 10% turbid fungal cell mass inoculum was prepared in 10 mL distilled water. Finally, fungal inoculum and Anthraquinonic (Basic Blue) dye was transferred into to 10 mL yeast peptone dextrose containing growth media in 45 mLsterile test tube and incubated in an incubator shaker at room temperature with agitation of 130 rpm for 45 days. Twenty-five fungal species decolorizing potential were determined using absorbance-transmittance percentage using BiOLOG turbidimeter. The efficiency of color removal (decolorization) was expressed as the percentage ratio of the decolorized dye concentration to that of initial one based on the following equation:
Anthraquinonic (Basic Blue) dye immersed in 10 mL sterile distilled water until it is adjusted to 0% Transmittance using BiOLOG turbidimeter and 10% turbid fungal cell mass inoculum was prepared in 10 mL distilled water. Finally, fungal inoculum and Anthraquinonic (Basic Blue) dye was transferred into to 10 mL yeast peptone dextrose containing growth media in 45 mLsterile test tube and incubated in an incubator shaker at room temperature with agitation of 130 rpm for 45 days. Twenty-five fungal species decolorizing potential were determined using absorbance-transmittance percentage using BiOLOG turbidimeter. The efficiency of color removal (decolorization) was expressed as the percentage ratio of the decolorized dye concentration to that of initial one based on the following equation:
Result
Screening and identification of species of fungi using BiOLOG MicroStaion
From waste damping site and rotten fruit 230 fungal isolates were screened and morphologically similar colonies Were clustered. Representative of clusters were transferred in to YT/FF MicroPlate for BiOLOG Micro Station reading. Finally, 35 fungal species (29 filamentous fungus and 7 yeast) were identified [Table1 and Table 2].
From waste damping site and rotten fruit 230 fungal isolates were screened and morphologically similar colonies Were clustered. Representative of clusters were transferred in to YT/FF MicroPlate for BiOLOG Micro Station reading. Finally, 35 fungal species (29 filamentous fungus and 7 yeast) were identified [Table1 and Table 2].
Decolorization/Removal%= Ci-Cf ×100 where (Ci) and (Cf) are the initial and final dye concentrations (mg/l), respectively
Ci
Ci
| No | List of identified fungal Species | Area isolated |
| 1 | Aspergillus carneus (V.Tiegham) Blockwitz | Adama |
| 2 | Penicillium digitatum (Pers.Fr) Sacc.BGA | Adama |
| 3 | Penicillium brevicompactum DierckxBGC | Adama/Gonder |
| 4 | Aspergillus ochraceus K.WilhelmBGB | Awoday |
| 5 | Aspergillus restrictus G.Sm | Awoday |
| 6 | Fusarium javanicum | Awoday |
| 7 | Penicillium roqueforti ThomBGE | Awoday |
| 8 | Aspergillus fresenii Subramaniam | Bahirdar |
| 9 | Aspergillus terricola var terricola | Bahirdar |
| 10 | Fusarium juruanum | Bahirdar |
| 11 | Penicillium solitum Westling BGA | Bahirdar |
| 12 | Cryptococcus albidus var albidus | Deredewa |
| 13 | Rhodotrula aurantica A | Deredwa |
| 14 | Candida glaebosa | Diredewa |
| 15 | Fellomyces fuzhouensis | Diredewa |
| 16 | Pichia norvegensis | Diredewa |
| 17 | Fusarium chlamydosporum var chlamydosporum wollenweber | Gonder |
| 18 | Cryptococcus lutelus | Harar/Diredewa/Jimma |
| 19 | Debermyomyces hansenii C | Jimma |
| 20 | Fusarium udume Butter | Jimma |
| 21 | Pencillium argillaceum stolk et al. | Jimma |
| 22 | Trichoderma aureovirida rafai | Jimma |
| 23 | Trichoderma piluiferum Webster & Rofi | Jimma |
| 24 | Trichosporon inkin | Jimma |
| 25 | Tricosporon begilli B | Jimma |
| 26 | Wingea robertsiae | Jimma |
| 27 | Pichia mexicana | Jimma/Diredewa |
| 28 | Pichia guilliermondii A | Mekele |
| 29 | Rhodotrula aurantica A | Mekele/Awoday/Jimma |
Table 1: Filamentous fungus species identified from Municipal solid waste.
| No | List of identified yeast Species | Area isolated |
| 1 | Candida apicola | Adama |
| 2 | Saccharomyces cerevisiae | Adama |
| 3 | Candida sorboxylosa | Adama/Gonder |
| 4 | Zygosaccaromycees fermentati | Awoday |
| 5 | Saccaromyces cerevisiae | Awoday |
| 6 | Isstachenkia orientalis | Awoday |
| 7 | Candida multisgmis | Awoday |
Table 2: Fungal species identified from rotten fruit.
Anthraquinonic (Basic Blue) dye degradation and decolorizing test using fungi
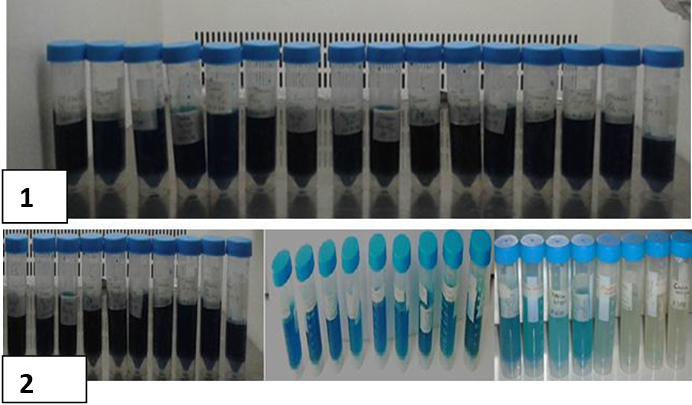
Twenty-five selected fungus species isolated from municipal solid waste and rotten fruit were tested for Anthraquinonic dye decolorization test based on absorbance and transmittance result of BiOLOG turbidimeter. Transmittance is simply the percentage of light impinging on a solution that passes through the solution and emerges to be detected by the instrument. It is zero for a completely opaque solution and 100% when all the light is transmitted. Four fungi strains, i.e., Candida multisgmis, Pichia guilliermonndi, Rhodotrula aurantica andTrichosporon begelli B showed 73-92% transmittance effective in decolorizing, four species, i.e., Hypopichia burtoni A, Rhodotrula aurentica B, Wingia robostam and Zygosaccaromycees fermentati showed 34-42% decolorizing ability. Nine species, i.e., Fusarium udum, Deberomyces hanseni, Pichia mexicana, kluyveromyces delphensis, Pencilium argillaceum, Pencilium rotifotrii, Pichia Mexicana and Trichoderma aureoviride showed 16-29% decolorizing ability. Nine fungal species, i.e., Candida sorboxylosa, Trichosporon piluilifera, Aspergillus restrictus,Candida apicola, Saccaromyces cerevisiae, Isstachenkia orientalis,Zygoascus hellensus, Fusarium chlymadospore and Pencillium digitatum did not have any decolorizing effect [Table 3; Figure 3 and Figure 4].
Twenty-five selected fungus species isolated from municipal solid waste and rotten fruit were tested for Anthraquinonic dye decolorization test based on absorbance and transmittance result of BiOLOG turbidimeter. Transmittance is simply the percentage of light impinging on a solution that passes through the solution and emerges to be detected by the instrument. It is zero for a completely opaque solution and 100% when all the light is transmitted. Four fungi strains, i.e., Candida multisgmis, Pichia guilliermonndi, Rhodotrula aurantica andTrichosporon begelli B showed 73-92% transmittance effective in decolorizing, four species, i.e., Hypopichia burtoni A, Rhodotrula aurentica B, Wingia robostam and Zygosaccaromycees fermentati showed 34-42% decolorizing ability. Nine species, i.e., Fusarium udum, Deberomyces hanseni, Pichia mexicana, kluyveromyces delphensis, Pencilium argillaceum, Pencilium rotifotrii, Pichia Mexicana and Trichoderma aureoviride showed 16-29% decolorizing ability. Nine fungal species, i.e., Candida sorboxylosa, Trichosporon piluilifera, Aspergillus restrictus,Candida apicola, Saccaromyces cerevisiae, Isstachenkia orientalis,Zygoascus hellensus, Fusarium chlymadospore and Pencillium digitatum did not have any decolorizing effect [Table 3; Figure 3 and Figure 4].
| S/N | Fungus species | Mean Percentage decolorization at 45 days |
| 1 | Candida multisgmis | 92% |
| 2 | Pichia guilliermonndi, | 87% |
| 3 | Rhodotrula aurantica A | 81% |
| 4 | Trichosporon begelli B | 73% |
| 5 | Hypopichia pichiaburtoni A | 42% |
| 6 | Rhodotrula aurentica B, | 37% |
| 7 | Wingia robosta | 36% |
| 8 | Zygosaccaromycees fermentati | 34% |
| 9 | Fusarium udum, | 29% |
| 10 | Deberomyces hanseni, | 26% |
| 11 | Pichia mexicana | 23% |
| 12 | kluyveromyces delphensis | 21% |
| 13 | Pencilium argillaceum | 19% |
| 14 | Pencilium rotifotrii | 18% |
| 15 | Pichia mexicana | 18% |
| 16 | Trichoderma aureoviride | 16% |
| 17 | Candida sorboxylosa | 0% |
| 18 | Trichosporon piluilifera | 0% |
| 19 | Aspergillus restrictus | 0% |
| 20 | Candida apicola | 0% |
| 21 | Saccaromyces cerevisiae | 0% |
| 22 | Isstachenkia orientalis | 0% |
| 23 | Zygoascus hellensus | 0% |
| 24 | Fusarium chlymadospore | 0% |
| 25 | Pencillium digitatum | 0% |
Table 3: Mean Percentage Decolorization of Anthraquinonic (Basic Blue) dye at 45 days incubation.
Discussion
The textile mills daily discharge millions of liters of untreated effluents in the form of wastewater into public drains that eventually empty into rivers. Most of them are recalcitrant in nature, especially Anthraquinonic (Basic Blue) dye. The stability and their xenobiotic nature of reactive dyes makes them recalcitrant hence they are not totally degraded by conventional wastewater treatment processes that involve light, chemicals or activated sludge (Chung., et al. 1992). The dyes are therefore released into the environment, in the form of colored wastewater. This can lead to acute effects on exposed organisms due to the toxicity of the dyes, phytoplankton’s form abnormal coloration and reduction in photosynthesis because of the absorbance of light that enters the water (Duran and Esposito 2000; Mester and Tien 2000).
This also alters the pH, increases the biochemical oxygen demand (BOD) and chemical oxygen demand (COD), and gives the rivers intense colorations and public is greatly concerned about water quality. The presence of unnatural colors is aesthetically unpleasant and tends to be associated with contamination. Without adequate treatment, these dyes will remain in the environment for an extended period of time (Olukanni., et al. 2006). Several physical, biological and chemical removal techniques like adsorption, coagulation, flocculation, membrane filtration, ozonation, electrochemical, radiolysis, advanced oxidation processes have been known to decolorize the textile effluents (Kannan and Sundaram, 2001; Rai., et al. 2005) which are very expensive and have drawbacks. Therefore, the advantage of using ecofriendly biological processes, such as the biotransformation of dyes, is that they may lead to complete mineralization or formation of less toxic products. The most widely researched fungi with regard to dye degradation are the ligninolytic fungi (Zille, 2005).
Different groups of fungi have been reported as producers of ligninolytic enzymes, but white-rot fungi have received extensive attention due to their powerful production of these enzymes and their unique ability to degrade lignin to CO2 (Lopez., et al. 2012). Such an extent of degradation is due to the strong oxidative activity and low substrate specificity of their ligninolytic system, which is primarily composed of laccase (Lac), Lignin peroxidase (LiP), and manganese peroxidase (MnP) (Martinez., et al. 2009). Anthraquinone dyes are used in textile, food, cosmetics, papermaking and pharmaceutical industries (Swamy and Ramsay, 1999; Padamavathy., et al. 2003). Their complex aromatic structure allows for their resistance to light and water oxidation and biodegradation. It is a feature desired by the industry using the dye, but it is dangerous for the environment. The non-biodegradable nature of most of the dyes causes their accumulation in surface water and sediments, especially at places of wastewater discharge. It reduces aquatic diversity by blocking the passage of sunlight through the water and creates problems for photosynthetic aquatic plants and algae, thus, affecting the ecological balance in the aquatic system. (Namasivayam and Sumithra, 2005; Divaniyam., et al. 2010). In this study 35 fungi identified with lactophenol cotton blue staining and BiOLOG Micro Staion identification system [Table1 and Table2].
Twenty five fungi were tested or evaluated for their degradation ability of Anthraquinonic dyes for 45 days in an aerobics condition, only four fungi species showed 73%-92% degree of transmittance measured using BiOLOG turbidimeter. 92% of transmittance recorded byCandida multisgmis, 87% ,81%,73% of transmittance recorded byPichia guilliermonndi, Rhodotrula aurantica A, and Trichosporon begelli B respectively [Table 3]. Atacag (2010) reported that Ascomyces yeast species such as Candida zeylanoides, Candida tropicalis, Debaryomyces polymorphus, Issatchenkia occidentalis, Candida albicans and Candida celeophila perform a putative enzymatic biodegradation and concomitant decolorization of several azodyes. Decolorization of an anthraquinone dye by white-rot fungi, Trametes versicolor and Phanerochaete chrysosporium, was investigated in a low-nitrogen and high-carbon liquid culture. Over 90% of the dye was decolorized after the 12h-incubation with T. versicolor. Very little work has been devoted to the study of the decolorizing ability of yeast, most often mentioning sorption as the main cause (Meehan., et al. 2000).
Dye decolorization by yeast generally occurs through three mechanisms, through bio sorption, bioaccumulation and biodegradation (Das and Charumathi, 2012).Galactomyces geotrichum, a yeast species, showed more than 96% decolorization of the azo dye, (Waghmode., et al. 2012). Abadulla (2000) reported that the anthraquinonic dyes decolorize faster than the azo dyes by Trametes hirsuta. Lokendra., et al. (2017) reported that 62% of degradation of anthraquinonic dye was recorded by Aspergillus flavus at 240 incubation hours. Wilkolazka., et al. (2002) reported that some brown fungi namely Coprinus micaccus, Fomtopsis pinicola, Gloephyllum odoratum, Aspergillus Niger, Aspergillus flavus, Aspergillus fumigatus. T. veride, F.oxysporum, P.chrysogenum, and Mucorspp. are responsible for degradation of anthraquinonic and wide range of textile dye. Shree (2015) reported that Bjekandera fumosa, Kuehneromyces mutabilis, Stropharia rugosoannulata resulted in great decolorization of basic blue dye up to 75-100%. The most widely explored fungi in regard to dye degradation are the ligninolytic fungi (Bumpus, 2004). Apart from this, Phanerochaete chrysosporium, Coriolus versicolar, Trametes versicolar, Fungalia trogii, Penicillium geastrivous, Rhizopus oryzar, Pleurotus ostreatus, Rigidoporus lignosus Pycnoporus sanguineus, Aspergillus flavus andAspergillus niger have been reported which are capable of degrading dyes (Fu and Viraraghavan, 2001; Wesenberg., et al. 2003).
White-rot fungi produce lignin peroxidase, manganese peroxidase and laccase that degrade many aromatic compounds due to their nonspecific enzyme systems. A strain of Candida zeylanoides has been successfully tested to degrade some dyes of simple chemical structure in liquid aerated bath culture. The amount of dye degradation varied from 43% to as high as 90% after 7 days. Remaxol Black B dye has been successfully decolorized using yeast Kluyveromyces marxianus. Maximum decolorization to the extent of 98% was observed at 37°C. Bacillus subtilis has also been used to degrade the structure of some dyes (Farida 2016). In this study four yeast species were capable of decolorizing the Anthraquinonic (Basic Blue) dye. HoweverCandida multisgmiswere superior in degradation and after further evaluation this would be a good candidate in biodegradation of Anthraquinonic (Basic Blue) dye and applied in textile waste water effluent treatment.
Conclusions
These results show that anthraquinone dyes are decolorized easier and faster by fungi. Among the 25 fungal strains tested, which had better potential for the dye decolorization, the strains such as Candida multisgmis Pichia guilliermonndi, Rhodotrula aurantica A, Trichosporon begelli B. as measured by the percentage of transmittance by turbidimeter. The strains of Candida multisgmis, Pichia guilliermonndi, Rhodotrula aurantica A were superior appear to be promising in terms of their Anthraquinonic (Basic Blue) dye color degradation that would be a good ecofriendly candidate in textile colored water effluent treatment after small scale evaluation.
Recommendation
The studies mainly cover chemically defined dyes, while the research with wastewater from dye stuff industry requires to be studied. Further research is also required in testing of these fungi species to other types of dyes. This research is at laboratory scale and small-scale evaluation is required at a textile plant.
The studies mainly cover chemically defined dyes, while the research with wastewater from dye stuff industry requires to be studied. Further research is also required in testing of these fungi species to other types of dyes. This research is at laboratory scale and small-scale evaluation is required at a textile plant.
References
- Abadulla E., et al. “Decolorization and detoxification of textile dyes with a laccase from Trameteshirsuta”. Applied Environmental Microbiology 66.8 (2000): 3357–3362.
- Atacag, EH., et al. “Biodegradation of Azo Dyes”. The Handbook of Environmental Chemistry 9 (2010): 201.
- Awaluddin R., et al. “Decolourization of commercial available synthetic dyes by the white-rot fungus, Phanerocheate chrysosporium ME446 (ATCC 4541)”. NSF workshop (2001), Kuala Lumpur, Malaysia.
- Baljeet SS and RangaP. “Optimization of cultural conditions for decolourization of textile azo dyes by Bacillus subtilis spr42 under submerged fermentation”. Advanced Biotechnology Research 2.1 (2011): 148–153.
- Bhatti HN., et al. “Optimization of culture conditions for enhanced decolorization of Cibacron Red FN-2BL by Schizophyllum commune IBL-6”. Applied Biochemistry and Biotechnology 149.3 (2008): 255–264.
- ChungK Tm and Cerniglia CE. “Mutagenicity of azo dyes: structure activity relationship”. Mutation Research 277.3 (1992): 201–220.
- Conneely A., et al. “Metabolism of the phthalocyanine textile dye remazol turquoise blue by Phanerochaete chrysosporium”. FEMS Microbiology Letters 179.2 (1999): 333–337.
- Cooper P. “Removing color from dye house wastewaters a critical review of technology available”. Journal of the Society of Dyers and Colorists 109.3 (1993): 97-100.
- Das N and Charumathi D. “Remediation of synthetic dyes from wastewater using yeast – An overview”. Indian Journal of Biotechnology 11 (2012): 369-380.
- Duran N and Esposito E. “Potential applications of oxidative enzymes and phenoloxidase-like compounds in wastewater and soil treatment: a review”. Applied Catalysis B: Environmental. 28.2 (2000): 83–99.
- Farida PM. “Microbial treatment of textile industry effluent: an ecofriendly recourse”. International Journal of Advanced Research 4.9 (2016): 1650-1654.
- Yuzhu., et al. “Removal of a dye from aqueous solution by the fungus Aspergillus niger”. Water QualityResearch Journal of Canada 35 (2002): 95-111.
- Gogate PR and Pandit AB. “A review of imperative technologies for waste water treatment II: Hybrid Methods”. Advances in Environmental Research 8. 3-4 (2004): 553-597.
- Kannan N and Sundaram MM. “Kinetics and mechanism of removal of methylene blue by adsorption on various carbons-a comparative study”. Dyes and Pigments51.1 (2001): 25-40.
- Katzbauer B., et al. “Classification system for immobilization techniques”. Bioprocess Engineering12.4 (1 995): 173–179.
- Lin JP., et al. “Production of Laccase by Coriolus Versicolor and Its Application in Decolorization of Dyestuffs: (II). Decolorization of Dyes by Laccase Containing Fermentation Broth With or Without Self-Immobilized Mycelia”. Journal of Environmental Sciences 1 5.1 (2003): 5-8.
- Lokendra S and Ved PS. “Decolourization of Azo (Acid Red) and Anthraquinonic (Basic Blue) Dyes by the Fungus Aspergillus flavus”. International Journal of Biomedical Engineering and Clinical Science 3.1 (2017): 1-5.
- Lopez JM., et al. “Pseudallescheria angusta, A LIGNINOLYTIC MICROORGANISM FOR WOOD FIBRES BIOMODIFICATION”. BioResources. 7.1 (2012): 464-474.
- Martinez AT., et al. “Enzymatic delignification of plant cell wall from nature to mill”. Current Opinion in Biotechnology 20.3 (2009): 348-357.
- Maximo C., et al. “Biotransformation of industrial reactive azo dye checked the ability of Pseudomonas sp. to remove colour by Geotrichum sp”. Enzyme and Microbial Technology 32 (2003): 145-151.
- Meehan CIM., et al. “Decolorization of Remazol Black-B using a thermo tolerant yeast, Kluyveromyces marxianus IMB3”. Environment International 26.1-2 (2000): 75-79.
- Mester T and Tien M. “Oxidative mechanism of ligninolytic enzymes involved in the degradation of environmental pollutants”. International Biodeterioration & Biodegradation 46.1 (2000): 51–59.
- Moorthi PS., et al. “Decolorization of textile dyes and their effluents using white rot fungi”. African Journal of Biotechnology 6.4 (2007): 424–429.
- Namasivayam C and Sumithra S. “Removal of direct red 12B and methylene blue from water by adsorption onto Fe (III)/Cr (III) hydroxide, an industrial solid waste”. Journal of Environmental Management74.3 (2005): 207–215.
- Olukanni OD., et al. “Textile effluent biodegradation potentials of textile effluent-adapted and non-adapted bacteria”. African Journal of Biotechnology 5.20 (2006): 1980-1984.
- O'Neill CFR., et al. “Colour in textile effluents-sources, measurement, discharge consents and simulation: A review”. Journal of Chemical Technology and Biotechnology 74.11 (1999): 1009-1018.
- Padamavathy S., et al. “Comparison of decolorization of reactive azo dyes by microorganisms isolated from various source”. Journal of Environmental Sciences 15.5 (2003): 628–632.
- Pointing SB and L.L.P.Vrijmoed. “Decolorization of azo and triphenylmethane dyes by Pycnoporus sanguineus producing laccase as the sole phenol oxidase”. World Journal of Microbiology and Biotechnology 16.3 (2000): 31 7–318.
- Puvaneswari N., et al.“Toxicity assessment and microbial degradation of azo dyes”. Indian Journal of Experimental Biology 44.8 (2006): 618-626.
- Rafi F., et al. “Optimization of cultural condition for decolorization of textile effluent”. Applied and Environmental Microbiology 56 (1990): 2146.
- Bhattacharyya MS., et al. “Removal of dyes from the effluent of textile and dyestuff manufacturing industry: A review of emerging techniques with reference to biological treatment”. Critical Reviews in Environmental Science and Technology 35.3 (2005): 219-238.
- Robinson T., et al. Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative”. Bioresource Technology 77.3 (2001):247–255.
- Rojek K., et al. “Decolorization of natural organic matter by Phanerochaete chrysosporium: The effect of environmental conditions”. Water Science and Technology: Water Supply 4.4 (2004): 1-75.
- Shree Nath Singh. “Microbial Degradation of Synthetic Dyes in Wastewaters”. Springer (2015): 215.
- Siddiqui MS., et al. “Up-flow immobilized fungal column reactor for the treatment of Anthraquinone dye Drimarene Blue K2RL”. African Journal of Biotechnology 8.20 (2009): 5570-5577.
- Siddiqui MS., et al. “Biotreatment of anthraquinone dye Drimarene Blue K2RL”. African Journal of Environmental Science and Technology 4.1 (2010): 045-050.
- Swamy J and Ramsay JA. “The evaluation of white-rot fungi in the decoloration of textile dyes”. Enzyme and Microbial Technology 24.3-4 (1999):130–137.
- Verma P and Madamwar D. “Decolorization of synthetic dyes by a newly isolated strain of Serratia marcescens”. World Journal of Microbiology and Biotechnology 19.6 (2003): 615–618.
- Waghmode RT., et al. “Degradation of Remazol Red dye by Galactomyces geotrichum MTCC 1360 leading to increased iron uptake in Sorghum vulgare and phaseolus mungo from soil”. Biotechnology and Bioprocess Engineering 17.1 (2012): 117-126.
- Wesenberg D., et al. “White-rot fungi and their enzymes for the treatment of industrial dye effluents”. Biotechnology Advances 22.1-2(2003): 161 –187.
- Zille A. “Laccase reactions for Textile Applications”. Thesis Submitted to Universidade do Minho (2005): 168.
- Zollinger H. “Colour chemistry synthesis-sis properties and application of organic dyes and pigments”. VCH New York: 92–102.ribution License (CC-BY) dyes and pigments.
Citation:
Genene Tefera., et al. “Fungi Isolated from Municipal Solid Waste Soil and Rotten Fruit Tested for their Ability to Decolorize
Anthraquinonic (Basic Blue) Dye”. Clinical Biotechnology and Microbiology 1.2 (2017): 82-92.
Copyright: © 2017 Genene Tefera., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.






























 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.