Research Article
Volume 1 Issue 4 - 2017
Bacterial Etiologies of Community-Acquired Pneumonia in Children and Their Antimicrobial Susceptibility Patterns
1Ethiopian Biodiversity Institute, Ethiopia
2Addis Ababa University, Faculty of Medicine, Department of Microbiology, Immunology and Parasitology
2Addis Ababa University, Faculty of Medicine, Department of Microbiology, Immunology and Parasitology
*Corresponding Author: Befekadu Teshome, Ethiopian Biodiversity Institute, Ethiopia.
Received: November 06, 2017; Published: November 13, 2017
Abstract
The study involved 205 children, aged 6 months to 12 years, with clinical signs of community-acquired pneumonia. During the period of June 2008 to January 2009, expectorated sputum samples were collected from Pediatric patients with community-acquired pneumonia who were attending the inpatient and outpatient departments at Tikur Anbassa University Hospital, Addis Ababa, Ethiopia.
Based on macroscopic and microscopic examination, 45 of 170 (25.8%) collected sputum specimens were determined to be suitable for culture. Sputum cultures were processed according to standard protocols. The isolated bacteria were identified by using API bacterial identification kits. The major pathogens for childhood community-acquired pneumonia were found to be S. pneumoniae, S. aureus, Acinetobacter spp., Moraxella spp., and Gram-negative enteric bacteria.
Antimicrobial susceptibility tests were done for the isolated pathogens by disk diffusion method. The S. pneumoniae isolates were 100% susceptible to penicillin, erythromycin, vancomycin, cefotaxime, ciprofloxacin, chloramphenicol, ampicillin and amoxicillin. There was 100% susceptibility of S. pyogenes to amoxicillin, chloramphenicol, ciprofloxacin, cefotaxime and clindamycin. There was also 100% susceptibility of Viridans Streptococci to amoxicillin, ampicillin, chloramphenicol, ciprofloxacin, clindamycin, and erythromycin. S. aureus showed high resistance to ampicillin (75%) and penicillin (87.5%).
The Gram-negative bacteria exhibited high resistance to ampicillin (97%), clindamycin (100%), erythromycin (87%), penicillin (100%) and vancomycin (100%). All Gram-negative bacteria were susceptible to ciprofloxacin and norfloxacin. Multi-drug resistance was observed in most of the isolated bacteria.
Keywords: Community-acquired pneumonia; Sputum; Bacterial etiology; Children
Introduction
Pneumonia is the result of infection and inflammation of the lung parenchyma distal to the terminal bronchioles (Black, 2008). The infection causes inflammation and deterioration of lung function. The lungs become unable to easily transfer oxygen to the blood, increasing the work of breathing. Pneumonia often affects only a portion of a single lung but can affect an entire lung or even both lungs. In many older people, the lung infection spreads beyond the lungs. The infection can enter the blood causing sepsis (Lutfiyya., et al. 2006).
In a patient with pre-existing respiratory disease; onset of bacterial pneumonia may result in deterioration of respiratory status, leading to respiratory failure and death (BTS, 2002). Pneumonia causes about 1 in 5 under five deaths worldwide: more than 2 million children each year. It kills more children than any other disease; more than AIDS, malaria and measles combined (UNICEF, 2006). In Africa, 21% of the 4.4 million deaths of children less than five years of age per year are caused by pneumonia The annual incidence of pneumonia in children younger than 5 years of age is 34 to 40 cases per 1000 in Europe and North America, higher than at any other time of life, except perhaps in adults older than 75 or 80 years of age.
In the developing world, pneumonia is not only more common than it is in Europe and North America; it is also more severe and is the largest killer of children (McIntosh, 2002). Pneumonia is the sixth leading cause of death in the United States of America and is the most common infectious cause of death. The mortality rate is reported to be 1% in the outpatient setting but may increase up to 25% in those requiring hospital admissions (Marrie, 1999).
Pneumonia is important in tropical and developing countries, due to its high lethality in children under 5 years old, especially among infants (March and Sant' Anna, 2005; Yoshimine., et al. 2001). In developed countries, this can be verified by the radiological finding of consolidation. In the developing world a more practical term-acute lower respiratory infection is preferred, reflecting the difficulties in obtaining a chest radiograph (BTS, 2002). The WHO (1994) has defined pneumonia solely on the basis of clinical findings obtained by visual inspection and timing of the respiratory rate.
Bacteria, the most common cause of CAP in children, traditionally have been divided into 2 groups; typical and atypical. Typical organisms include Streptococcus pneumoniae and Haemophilus and Staphylococcus species. Atypical refers to pneumonia caused by Legionela, Mycoplasma and Chlamydia (Marrie, 1999). The focus of this study was on the identification of the typical bacteria. Identification of the etiologic agent of CAP has theoretically important advantages.
For individual patients, it facilitates the administration of a targeted, narrow-spectrum antibiotic therapy that may improve the efficacy of treatment, and reduces risks of antibiotic related toxicity. For the community, microbiologic results contribute to understanding of the local microbial epidemiology of CAP and the antimicrobial resistance patterns of pathogens. This information is essential in the instauration or modification of empiric treatment regimens. To save lives and reduce mortality with proper treatment, there is no alternative other than to obtain this information for each country (Ozyilmaz., et al. 2005).
As most cases of severe pneumonia are caused by bacterial pathogens, prompt treatment with effective antibiotics is life-saving. Knowing the pathogens that lead to pneumonia is critical for guiding antibiotic treatment and policies. Information in this regard is scarce in Ethiopia. Thus, the purpose of this study was to determine bacterial Etiologies of CAP in children and their resistance patterns to antibiotics, which can be useful in the general treatment option of CAP.
Materials and Methods
Study design
A hospital based cross-sectional study was conducted at the Pediatric OPD in Tikur Anbassa Hospital, Addis Ababa, Ethiopia from June 2008 through January 2009. The study was reviewed and approved by the department of Microbiology, Immunology, and Parasitology ethical committee and Institutional Review Board of the Medical Faculty of Addis Ababa University. Signed informed parental consent and the child's assent (if the child was < 10 years old) were obtained. All the information obtained was kept confidential. If patients were not interested in the study, they had the right to withdraw from the study.
A hospital based cross-sectional study was conducted at the Pediatric OPD in Tikur Anbassa Hospital, Addis Ababa, Ethiopia from June 2008 through January 2009. The study was reviewed and approved by the department of Microbiology, Immunology, and Parasitology ethical committee and Institutional Review Board of the Medical Faculty of Addis Ababa University. Signed informed parental consent and the child's assent (if the child was < 10 years old) were obtained. All the information obtained was kept confidential. If patients were not interested in the study, they had the right to withdraw from the study.
Study populations
Patients were recruited in the paediatric outpatient department of Tikur Anbassa Hospital, Addis Ababa, Ethiopia. The samples were collected from children aged 6 months to 12 years with suspected CAP. The population at large is served in this health facility that comes from the city of Addis Ababa and different parts of Ethiopia. Two hundred five children with CAP were included in the study. All children aged 6 months to 12 years admitted to Pediatric OPD emergency room with suspect community-acquired pneumonia were included in the study.
Patients were recruited in the paediatric outpatient department of Tikur Anbassa Hospital, Addis Ababa, Ethiopia. The samples were collected from children aged 6 months to 12 years with suspected CAP. The population at large is served in this health facility that comes from the city of Addis Ababa and different parts of Ethiopia. Two hundred five children with CAP were included in the study. All children aged 6 months to 12 years admitted to Pediatric OPD emergency room with suspect community-acquired pneumonia were included in the study.
Eligibility and exclusion criteria
Community-acquired pneumonia was defined as pneumonia that has been acquired in the community in a patient who has not been hospitalized within 14 days prior to onset of symptoms or has been hospitalized for less than 4 days prior to the onset of symptoms. Patients eligible for inclusion in the study met the following criteria:
Community-acquired pneumonia was defined as pneumonia that has been acquired in the community in a patient who has not been hospitalized within 14 days prior to onset of symptoms or has been hospitalized for less than 4 days prior to the onset of symptoms. Patients eligible for inclusion in the study met the following criteria:
- Children aged 6 months to 12 years with a primary (putative) diagnosis of CAP made within 24 hrs of admission; a medical and clinical history of pneumonia
- Clinical observation of two or more signs and symptoms associated with a lower respiratory tract infection (i.e. body temperature >37.8°C, new or increased cough, production of purulent sputum) or at least two of the minor conditions, which include: pleuritic chest pains, dyspnoea, altered mental status, pulmonary consolidation by physical examination.
- Children with CAP who have been hospitalized less than 4 days prior to the onset of symptoms.
- Patients in the following categories were excluded from the study: children < 6 months of age; and admission from a nursing home or a hospital within a month prior to the study so as to avoid the inclusion of possible nosocomial case.
Sample collection and transport
Within 24 hrs of admission, a physician examined the patients and recorded the findings on standardized case record forms. Nurse collected samples of sputum from children with suspected CAP in sterile, screw top sputum cups. The expectorated sputum was collected, ideally a minimum volume of 1 ml. by asking the patient to cough deeply into the container, followed by immediate screwing on the cup.
Within 24 hrs of admission, a physician examined the patients and recorded the findings on standardized case record forms. Nurse collected samples of sputum from children with suspected CAP in sterile, screw top sputum cups. The expectorated sputum was collected, ideally a minimum volume of 1 ml. by asking the patient to cough deeply into the container, followed by immediate screwing on the cup.
No special procedures were performed to obtain sputum samples if they could not be obtained spontaneously. Samples were transported to the laboratory within two hrs of collection and processed immediately as explained by Yamazaki., et al. (2005) and Nagalingam., et al. (2005). Sample processing and Microbiological examination was done at Bacteriologic Unit of Tikur Anbassa University Hospital.
Microbiological examinations
Microscopy: Sputum Gram stains
A smear was prepared from the most purulent materials in the sputum and a Gram stain was done using standard methods (Bauer., et al. 1966). The quality of the sputum specimen was assessed by evaluating the Gram stain slide under light microscope (X10). Polymorpho nuclear leucocytes and squamous epithelial cells were counted.
Microscopy: Sputum Gram stains
A smear was prepared from the most purulent materials in the sputum and a Gram stain was done using standard methods (Bauer., et al. 1966). The quality of the sputum specimen was assessed by evaluating the Gram stain slide under light microscope (X10). Polymorpho nuclear leucocytes and squamous epithelial cells were counted.
Sputum samples were considered of good quality if they had less than 10 squamous epithelial cells and more than 25 Polymorphonuclear neutrophil per low power field. Otherwise, the sputum sample was considered contaminated by saliva and discarded (Garcia-Vazquez., et al. 2004; Roson., et al. 2000; Nagalingam., et al. 2005).
Good quality specimens were then screened for a predominant bacterial morphological type at oil immersion field (X100). Quantitation of these parameters was accomplished using the following criteria: (0, none seen; 1+, 1-5; 2+, 5-10; 3+, 11-25; and 4+, > 25 per field) (Nagendra., et al. 2001). A predominant morphotype was defined as the presence of a single morphotype that accounted for > 75% of the organism seen (Roson., et al. 2000).
Isolation and Identification of bacterial pathogens
A proportion of another purulent area of the good quality sputum was used for microbiological analysis. Sputum cultures were processed immediately in blood agar, chocolate agar, and MacConkey agar media. The following organisms were considered potential pathogens: S. pneumoniae, H. influenzae, M. catarrhalis, S. aureus, beta-haemolytic streptococci, Enterobacteriaceae, Pseudomonas and Acinetobacter sp. The organism was considered as the potential etiology based on the definition of Fang., et al. (1990).
A proportion of another purulent area of the good quality sputum was used for microbiological analysis. Sputum cultures were processed immediately in blood agar, chocolate agar, and MacConkey agar media. The following organisms were considered potential pathogens: S. pneumoniae, H. influenzae, M. catarrhalis, S. aureus, beta-haemolytic streptococci, Enterobacteriaceae, Pseudomonas and Acinetobacter sp. The organism was considered as the potential etiology based on the definition of Fang., et al. (1990).
Five percent sheep blood agar was used to isolate and identify Gram-positive organisms. The plates were incubated in a 5-10% candle extinction jar at 35-37OC for 18-24 hrs. On the next day, identification of colonies, Gram staining, and sub-culturing were done. The potential Gram-positive diplococci and Gram-positive cocci pathogens of CAP were identified based on the standard microbiological and biochemical techniques (NCCLS, 2001; Bergey’s Manual of systematic Bacteriology, 2010).
Chocolate agar was used to isolate and identify H. influenzae and M. catarrhalis. Sputum sample was inoculated to CA by taking a loopful of specimen. The plates were incubated at 35-37OC in 5-10% CO2 for 18-24 hrs (Nagalingam., et al. 2005; Bergey’s Manual of systematic Bacteriology, 2010). Confirmation was made by API 10NH system (BioMerieux, France) according to the manufacturer¢s standard (Guclu., et al. 2005) and by demonstrating satillitism with the x and v test.
GNEB were isolated from the sputum sample by plating onto a MacConkey agar plate, incubated at 35-37OC aerobically for 24 hrs. GNEB and Pseudomonas were identified based on the standard microbiological and biochemical techniques (Shah., et al. 2002). Confirmation was made by API 20E system (BioMerieux, France) according to the manufacturer¢s standard (Guclu., et al. 2005).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was measured using the agar disk diffusion method as outlined by Bauer., et al. (1966). Mueller Hinton Agar was used for susceptibility testing of the isolated bacteria. The Mueller Hinton Agar medium was supplemented with 5% sheep blood for susceptibility testing of S. pneumoniae and other Streptococci species (NCCLS, 2001).
Antimicrobial susceptibility was measured using the agar disk diffusion method as outlined by Bauer., et al. (1966). Mueller Hinton Agar was used for susceptibility testing of the isolated bacteria. The Mueller Hinton Agar medium was supplemented with 5% sheep blood for susceptibility testing of S. pneumoniae and other Streptococci species (NCCLS, 2001).
Generally, 13 antibiotic disks (Oxoid, Basingstoke, Hampshire, England) were tested: amoxicillin (30 mg), ampicillin (10 mg), gentamycin (10 mg), erythromycin (15 mg), ciprofloxacin (5 mg), chloramphenicol (30 m), cefotaxime (10 mg), vancomycin (2 mg), trimethoprim-sulfamethoxazole (25 mg), penicillin G (10 mg), tetracycline (30 mg), clindamycin (2 mg), and norfloxacin (10 mg).
The diameter of each zone of inhibition was measured and interpretation of susceptibility was done by comparing the results to the standard zone sizes recommended by NCCLS (2001) and labelled as resistant, intermediate and susceptible.
Standard control reference strains: The reference control organisms used were Staphylococcus aureus (ATCC 25923), Pseudomonas aeruginosa (ATCC 27833) and Escherichia coli (ATCC 25922) which were obtained from EHNRI. All in vitro tests were done strictly according to the research protocol, and a 4-day quality control run-in period was followed by actual in vitro studies.
Statistical analysis of data
The data obtained from this study were entered into a computer and all statistical values were analysed using SPSS soft-ware package, version 15. Descriptive data were presented as means ± SD for continuous variables and as rates for categorical variables. Statistical comparisons of categorical variables were made by Chi-square analysis or the Fisher exact test, when appropriate. Statistical significance was defined as P < 0.05 (2-tailed).
The data obtained from this study were entered into a computer and all statistical values were analysed using SPSS soft-ware package, version 15. Descriptive data were presented as means ± SD for continuous variables and as rates for categorical variables. Statistical comparisons of categorical variables were made by Chi-square analysis or the Fisher exact test, when appropriate. Statistical significance was defined as P < 0.05 (2-tailed).
Results
Patient demographics
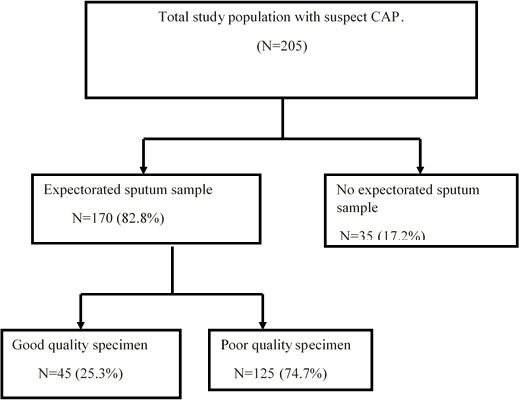
A total of 205 children with community-acquired pneumonia met the inclusion criteria for enrolment (Figure 1). Eleven cases of the 205 patients were radiographically confirmed to have community-acquired pneumonia, as diagnosed by radiologists in Tikur Anbassa Hospital. The diagnosis of lower respiratory involvement was based on clinical signs and symptoms which were made by the attending physicians in Pediatric OPD. Of these patients, 98 (47.8%) were males. Ages ranged from 6 months to 12 years with mean of 5.13 years.
A total of 205 children with community-acquired pneumonia met the inclusion criteria for enrolment (Figure 1). Eleven cases of the 205 patients were radiographically confirmed to have community-acquired pneumonia, as diagnosed by radiologists in Tikur Anbassa Hospital. The diagnosis of lower respiratory involvement was based on clinical signs and symptoms which were made by the attending physicians in Pediatric OPD. Of these patients, 98 (47.8%) were males. Ages ranged from 6 months to 12 years with mean of 5.13 years.
Seventy-three (36.0%) were 6 months to < 2 years old, 37 (18.0%) were 2 to < 5 years old, and 95 (46.0%) were ³ 5 years to 12 years old. Most had been ill for less than two weeks. The mean duration of illness prior to coming to hospital was 7.9 days. One hundred and thirty-three (64.9%) patients had received antibiotics prior to admission.
The most common antibiotics used were penicillin 63 (47.4%), ceftriaxon 31 (23.3%), ampicillin 16 (12.0%), amoxicillin and gentamycin 15 (11.3%) and chloramphenicol 8 (6.0%). The most common symptoms and signs which were observed in the patients were: cough 163 (79.5%), difficulty breathing 127 (62.0%), fever 123 (60.0%), chest pains 10 (4.9%) and convulsion 9 (4.4%).
Sputum examination result: Macroscopic and Microscopic
Sputum samples were obtained from 170 patients (82.3% of the total) (Figure 1). By macroscopic examination at the appearance of the 170 collected sputum specimens, 101 (59.4%) were found to be serous, 49 (28.8%) purulent, 14 (8.2%) mucoid and 6 (3.5%) seropurulent. Concerning their color, 15 (8.8%) of the samples were blood-streaked sputum, 54 (31.5%) yellow-green and 101 (59.4%) clear and colourless. One hundred and sixty-nine (99.4%) of them did not have putrid odor. By observing the status of PMNs and the epithelial cells in a Gram-stained smear, 45 of 170 (25.3%) collected sputum specimens were determined to be suitable for culture. Thus, the final study populations consisted of 45 children from whom good quality sputum samples were obtained (Figure 1).
Sputum samples were obtained from 170 patients (82.3% of the total) (Figure 1). By macroscopic examination at the appearance of the 170 collected sputum specimens, 101 (59.4%) were found to be serous, 49 (28.8%) purulent, 14 (8.2%) mucoid and 6 (3.5%) seropurulent. Concerning their color, 15 (8.8%) of the samples were blood-streaked sputum, 54 (31.5%) yellow-green and 101 (59.4%) clear and colourless. One hundred and sixty-nine (99.4%) of them did not have putrid odor. By observing the status of PMNs and the epithelial cells in a Gram-stained smear, 45 of 170 (25.3%) collected sputum specimens were determined to be suitable for culture. Thus, the final study populations consisted of 45 children from whom good quality sputum samples were obtained (Figure 1).
Thirteen of the 45 good-quality samples (28.9%) showed PMs on sputum Gram stains. Eight of them (61.5%) were Gram-positive diplococci, 3 of them (23.1%) Gram-positive cocci and 2 of them (15.4%) were Gram-negative bacilli (Table 1). Results for patients with previous antibiotic treatment are also shown in Table 1.
| Gram stain result | Patients who provided sputum (N = 170) |
Patients with PAT and who provided sputum (N = 119) |
| n (%) | n (%) | |
| Good quality samples | 45 (25.3) | 31 (26.1) |
| Predominant morphotype | 13 (28.9) | 6 (19.4) |
| Gram-positive diplococci | 8 (61.5) | 3 (50.0) |
| Gram-positive cocci | 3 (23.1) | 2 (33.3) |
| Gram-negative bacilli | 2 (15.4) | 1 (16.7) |
Table 1: Bacterial distribution in sputum Gram stains of 170 patients with CAP.
CAP: Community-acquired pneumonia, PAT: Previous antibiotic treatment. Statistical significance was defined as P < 0.05
CAP: Community-acquired pneumonia, PAT: Previous antibiotic treatment. Statistical significance was defined as P < 0.05
Etiologic agents
The bacteria isolated from sputum collected for this study are shown in Tables 2 and Tables 3. Evidence of bacterial infection was detected in 36 of the 45 (80.0%) children who provided good quality sputum samples. A single bacterial infection was found in 17 of the 45 (38.0%) children. Two or more bacteria were found in 19 of the 45 (42.0%) children and no pathogen was found in 9 of the 45 (20.0%) sputum cultures (Table 2).
The bacteria isolated from sputum collected for this study are shown in Tables 2 and Tables 3. Evidence of bacterial infection was detected in 36 of the 45 (80.0%) children who provided good quality sputum samples. A single bacterial infection was found in 17 of the 45 (38.0%) children. Two or more bacteria were found in 19 of the 45 (42.0%) children and no pathogen was found in 9 of the 45 (20.0%) sputum cultures (Table 2).
Sixty-three isolates were cultured from 36 patients with community-acquired pneumonia. Overall, 32 of 63 (50.8%) were Gram-positive organisms and 31 of 63 (49.2%) were Gram-negative bacteria. S. pneumoniae and S. pyogenes, each accounted for 14.3% of the total isolated bacteria, followed by S. aureus (12.5%), E. cloacae (11.1%), Viridans Streptococci (9.5%), Acinetobacter spp (7.9%), E. coli, S. marcescens and Klebsiella spp (K. pneumoniae and K. oxytoca) (6.3%), M. catarrhalis (4.8%), and P. aeruginosa, P. mirabilis, C. freundi and Aeromonas spp. (1.6%) (Table 3)
| Isolated bacteria | No. of cases (%) (N = 45) |
| Single pathogen | 17 (38.0) |
| S. pneumoniae | 5 (29.4) |
| S. aureus | 4 (23.5) |
| Viridans Streptococci | 3 (17.6) |
| S. pyogenes | 2 (11.8) |
| K. pneumoniae | 1 (5.9) |
| E. cloacae | 1 (5.9) |
| S. marcescens | 1 (5.9) |
| Multiple pathogens | 19 (42.0) |
| S. pyogenes + E. coli | 2 (10.5) |
| S. pneumoniae + P. aeruginosa | 1 (5.3) |
| S. pneumoniae + S. pyogenes | 1 (5.3) |
| S. pneumoniae + Viridans Streptococci | 1 (5.3) |
| S. pneumoniae + S. marcescens | 1 (5.3) |
| S. pyogenes + Viridans Streptococci | 1 (5.3) |
| S. pyogenes + Aeromonas spp | 1 (5.3) |
| S. aureus + E. coli | 1 (5.3) |
| M. catarrhalis + K. oxytoca | 1 (5.3) |
| E. cloacae + Acinetobacter spp | 1 (5.3) |
| S. aureus + M. catarrhalis + E. cloacae | 1 (5.3) |
| S. aureus + P. mirabilis + Viridans Streptococci | 1 (5.3) |
| S. aureus + E. coli + Acinetobacter spp | 1 (5.3) |
| S. pyogenes + E. cloacae + Acinetobacter spp | 1 (5.3) |
| S. pyogenes + K. oxytoca + Acinetobacter spp | 1 (5.3) |
| E. cloacae + S. marcescens + Acinetobacter spp | 1 (5.3) |
| E. cloacae + K. pneumoniae + S. marcescens | 1 (5.3) |
| E. cloacae + M. catarrhalis + C. freundi | 1 (5.3) |
Table 2: Identified causative bacteria from 45 expectorated sputa cultures of cases with CAP.
9 of the 45 cultures were negative (i.e. 20.0%)
9 of the 45 cultures were negative (i.e. 20.0%)
| Characteristics | Patients who provided sputum (N = 170) |
Patients with PAT and who provided sputum (N = 119) |
| n (%) | n (%) | |
| Gram positive bacteria | 32 (50.8) | 19 (40.4) |
| S. pneumoniae | 9 (14.3) | 5 (10.6) |
| S. pyogenes | 9 (14.3) | 5 (10.6) |
| Viridans Streptococci | 6 (9.5) | 4 (8.5) |
| S. aureus | 8 (12.5) | 5 (10.6) |
| Gram negative bacteria | 31 (49.2) | 28 (59.6) |
| Moraxella catarrhalis | 3 (4.8) | 3 (6.4) |
| Acinetobacter spp | 5 (7.9) | 5 (10.6) |
| E. cloacae | 7 (11.1) | 6 (12.8) |
| E. coli | 4 (6.3) | 3 (6.4) |
| S. marcescens | 4 (6.3) | 4 (8.5) |
| K. pneumoniae | 2 (3.2) | 2 (4.3) |
| K. oxytoca | 2 (3.2) | 2 (4.3) |
| P. aeruginosa | 1 (1.6) | 0 (0) |
| P. mirabilis | 1 (1.6) | 1 (2.1) |
| C. freundi | 1 (1.6) | 1 (2.1) |
| Aeromonas spp | 1 (1.6) | 1 (2.1) |
| Total | 63 (100.0) | 47 (100.0) |
Table 3: Pathogen distribution in the sputum samples of 170 patients with CAP.
CAP: community-acquired pneumonia, PAT: Previous antibiotic treatment, Statistical significance was defined as P < 0.05
CAP: community-acquired pneumonia, PAT: Previous antibiotic treatment, Statistical significance was defined as P < 0.05
Antimicrobial susceptibility test results
The antibiotic susceptibility patterns of the isolated bacteria from the sputum samples for Gram-positive and Gram-negative bacteria are shown in Tables 4 and 5. All S. pneumoniae isolates were susceptible to penicillin, erythromycin, vancomycin, cefotaxime, ciprofloxacin, chloramphenicol, ampicillin and amoxicillin. The results of susceptibility testing of S. pneumoniae to other antimicrobial agents revealed the following resistance rates: tetracycline (33.3%), trimethoprim-sulphamethoxazole (33.3%), norfloxacin (22.2%), clindamycin (11.1%) and gentamycin (11.1%). All S. pyogenes isolates were susceptible to amoxicillin, chloramphenicol, ciprofloxacin, cefotaxime and clindamycin. All isolates of Viridans streptococci were susceptible to amoxicillin, ampicillin, chloramphenicol, ciprofloxacin, clindamycin, and erythromycin. S. aureus showed high resistance to ampicillin (75.0%) and penicillin (87.5%). S. aureus showed no resistance to ciprofloxacin, vancomycin and norfloxacin. The Gram-negative bacteria exhibited high level of resistance to ampicillin (97.0%), clindamycin (100.0%), erythromycin (87.0%), penicillin (100.0%) and vancomycin (100.0%). All Gram-negative bacteria were susceptible to ciprofloxacin and norfloxacin except one strain of Acinetobacter sp.
The antibiotic susceptibility patterns of the isolated bacteria from the sputum samples for Gram-positive and Gram-negative bacteria are shown in Tables 4 and 5. All S. pneumoniae isolates were susceptible to penicillin, erythromycin, vancomycin, cefotaxime, ciprofloxacin, chloramphenicol, ampicillin and amoxicillin. The results of susceptibility testing of S. pneumoniae to other antimicrobial agents revealed the following resistance rates: tetracycline (33.3%), trimethoprim-sulphamethoxazole (33.3%), norfloxacin (22.2%), clindamycin (11.1%) and gentamycin (11.1%). All S. pyogenes isolates were susceptible to amoxicillin, chloramphenicol, ciprofloxacin, cefotaxime and clindamycin. All isolates of Viridans streptococci were susceptible to amoxicillin, ampicillin, chloramphenicol, ciprofloxacin, clindamycin, and erythromycin. S. aureus showed high resistance to ampicillin (75.0%) and penicillin (87.5%). S. aureus showed no resistance to ciprofloxacin, vancomycin and norfloxacin. The Gram-negative bacteria exhibited high level of resistance to ampicillin (97.0%), clindamycin (100.0%), erythromycin (87.0%), penicillin (100.0%) and vancomycin (100.0%). All Gram-negative bacteria were susceptible to ciprofloxacin and norfloxacin except one strain of Acinetobacter sp.
| Antimicrobial agents | S. pneumoniae N = 9 |
S. pyogenes N = 9 |
S. aureus N = 8 |
Viridans Streptococci N = 6 |
| No. (%) | No. (%) | No. (%) | No. (%) | |
| AMC (30 µg) | 0 (0.0) | 0 (0.0) | 3 (37.5) | 0 (0.0) |
| AMP (10 µg) | 0 (0.0) | 1 (11.1) | 6 (75.0) | 0 (0.0) |
| C (30 µg) | 0 (0.0) | 0 (0.0) | 2 (25.0) | 0 (0.0) |
| CIP (5 µg) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| CN (10 µg) | 1 (11.1) | 3 (33.3) | 2 (25.0) | 2 (33.3) |
| CTX (10 µg) | 0 (0.0) | 0 (0.0) | 5 (62.5) | 2 (33.3) |
| DA (2 µg) | 1 (11.1) | 0 (0.0) | 1 (12.5) | 0 (0.0) |
| E (15 µg) | 0 (0.0) | 1 (11.1) | 1 (12.5) | 0 (0.0) |
| NOR (10 µg) | 2 (22.2) | 3 (33.3) | 0 (0.0) | 3 (50.0) |
| P (10 µg) | 0 (0.0) | 1 (11.1) | 7 (87.5) | 1 (16.7) |
| SXT (25 µg) | 3 (33.3) | 2 (22.2) | 1 (12.5) | 4 (66.7) |
| TE (30 µg) | 3 (33.3) | 4 (44.4) | 5 (62.5) | 2 (33.3) |
| VA (2 µg) | 0 (0.0) | 1 (11.1) | 0 (0.0) | 3 (50.0) |
Table 4: Resistant percentage of the isolated Gram-positive bacteria to selected antimicrobial agents.
AMC: Amoxicillin-Clavulanic acid; AMP: Ampicillin; CN: Gentamycin; CIP: Ciprofloxacin; C: Chloramphenicol; CTX: Cefotaxime; DA: Clindamycin; E: Erythromycin; NOR: Norfloxacin; P: Penicillin; SXT: Trimethoprim-sulphamethoxazole; TE: Tetracycline; VA: Vancomycin
AMC: Amoxicillin-Clavulanic acid; AMP: Ampicillin; CN: Gentamycin; CIP: Ciprofloxacin; C: Chloramphenicol; CTX: Cefotaxime; DA: Clindamycin; E: Erythromycin; NOR: Norfloxacin; P: Penicillin; SXT: Trimethoprim-sulphamethoxazole; TE: Tetracycline; VA: Vancomycin
| Antimicrobial agents | M. catarrhalis n = 3 No. (%) |
Acinetobacter spp n = 5 No. (%) |
E. cloacae n = 7 No. (%) |
E. coli n = 4 No. (%) |
S. marcescens n = 4 No. (%) |
K. pneumoniae n = 2 No. (%) |
K. oxytoca n = 2 No. (%) |
P. aeruginosa n = 1 No. (%) |
P. mirabilis n = 1 No. (%) |
C. freundi n = 1 No. (%) |
Aeromonas spp n = 1 No. (%) |
| AMC (30 µg) | 3 (100.0) | 5 (100.0) | 4 (57.1) | 3 (75.0) | 2 (50.0) | 1(50.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 1 (100.0) |
| AMP (10 µg) | 3 (100.0) | 5 (100.0) | 6 (85.7) | 4 (100.0) | 4 (100.0) | 2 (100.0) | 2 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| C (30 µg) | 0 (0.0) | 3 (60.0) | 1(14.3) | 3(75.0) | 0 (0.0) | 1(50.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) |
| CIP (5µg) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| CN (10 µg) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (25.0) | 0 (0.0) | 1 (50.0) | 2 (100.0) | 0 (0.0) | 0(0.0) | 0 (0.0) | 0 (0.0) |
| CTX (10 µg) | 3 (100.0) | 5 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| DA (2 µg) | 3 (100.0) | 5 (100.0) | 7 (100.0) | 4 (100.0) | 4 (100.0) | 2 (100.0) | 2 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| E (15 µg) | 1 (33.3) | 3 (60.0) | 7 (100.0) | 4 (100.0) | 4 (100.0) | 2 (100.0) | 2 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| NOR (10 µg) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| P (10 µg) | 3 (100.0) | 4 (80.0) | 7 (100.0) | 4 (100.0) | 4 (100.0) | 2 (100.0) | 2 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| SXT (25 µg) | 0 (0.0) | 1 (20.0) | 5 (71.4) | 3 (75.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
| TE (30 µg) | 0(0.0) | 2 (40.0) | 2 (28.6) | 2 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) |
| VA (2 µg) | 3 (100.0) | 5 (100.0) | 7 (100.0) | 4 (100.0) | 4 (100.0) | 2 (100.0) | 2 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) | 1 (100.0) |
Table 5: Resistance percentage of the isolated Gram-negative bacteria to selected antimicrobial agents.
AMC: Amoxicillin; AMP: Ampicillin; C: Chloramphenicol; CIP: Ciprofloxacin; CN: Gentamycin; CTX; Cefotaxime; DA: Clindamycin; E: Erythromycin; NOR: Norfloxacin; P: Penicillin; SXT: Trimethoprim-sulphamethoxazole; TE: Tetracyclin; VA: Vancomycin;
AMC: Amoxicillin; AMP: Ampicillin; C: Chloramphenicol; CIP: Ciprofloxacin; CN: Gentamycin; CTX; Cefotaxime; DA: Clindamycin; E: Erythromycin; NOR: Norfloxacin; P: Penicillin; SXT: Trimethoprim-sulphamethoxazole; TE: Tetracyclin; VA: Vancomycin;
Discussion
By observing the status of Polymorphonuclear neutrophils and the epithelial cells in a Gram-stained smear, 45 of 170 (25.3%) collected sputum specimens were determined to be suitable for culture. From the 45 sputum specimens which were determined to be suitable for culture, 36 of them (80.0%) found to be culture-positive. The most common pathogen was S. pneumoniae, which was isolated from 9 patients. The finding of 9 S. pneumoniae isolates represents 14.3% of the total isolated bacteria.
This result is related to the 8% prevalence of S. pneumoniae in HIV-uninfected persons in Uganda (Yoshimine., et al. 2001), 6% prevalence of S. pneumoniae in Addis Ababa (Aderaye, 1994) and 18% in Zimbabwe (Nascimento-Carvalho, 2001). Other studies have revealed comparable values, as was found in Turkey, where an isolation rate of 25.5% was found amongst 98 patients (Ozyilmaz., et al. 2005). However, the detected S. pneumoniae from sputum of the pneumonia cases in this study is less frequent than in some published studies where other methods of detection were used.
Bacterial findings in studies from Africa by using lung aspirates showed prevalence of 35% in Nigerian children and 51% in Gambian children for S. pneumoniae (Nascimento-Carvalho, 2001). Since most studies employed 2 or 3 different diagnostic methods in combination to find this bacterium, caution must therefore be taken when comparing prevalence data on S. pneumoniae. This is a low yield as is the case in most developing countries where culture facilities are either unavailable or grossly inadequate. Besides, 64.9% of the population included in the study had taken antibiotics prior to coming to hospital and this might have compounded the problem.
The zero prevalence (0.0%) of H. influenzae found in community-acquired pneumonia cases in this study is similar to published reports, such as 0.8% by Nagalingam et al. (2005), and 0% by Bochud., et al. (2001) in Switzerland. Michelow., et al. (2004) included 154 children in their study and used blood or pleural fluid cultures, pneumolysin-based polymerase chain reaction assays and serologic tests to clarify the epidemiology but H. influenzae was not found to be a causative pathogen of community-acquired pneumonia in their study. Since most patients who were included in this study did take antibiotic treatment before collection of sputum, due to this reason the isolation of H. influenzae from the specimens might become nil.
The prevalence of 12.5% was found for S. aureus. This prevalence of S. aureus is similar to the 12.0% found by Lahti., et al. (2009). Bacteriologic findings of 9% prevalence of S. aureus in Nigeria and 10% prevalence of S. aureus in Zimbabwe (Nascimento-Carvalho, 2001) were found which are comparable to the findings of this study.
The prevalence of 4.8% for Moraxella catarrhalis in the present study is similar to the 6% prevalence of etiology of childhood community-acquired pneumonia in Europe using non-invasive diagnostic techniques (Nascimento-Carvalho, 2001). Ozyilmaz., et al. (2005) found 12.2% prevalence of Moraxella catarrhalis in Turkey. Lahti., et al. (2009) also found 28% prevalence by using induced sputum. Moraxella catarrhalis is considered as a less common agent of community-acquired pneumonia (Bartlett and Mundy, 1995).
Reported prevalence of community-acquired pneumonia in the general population due to Gram-negative enteric bacteria varies from 0% to 9% (Nagalingam., et al. 2005). The prevalence of 11.1% for Enterobacter cloacae in the present study is comparable to the isolation of 17.8% prevalence reported by Buenviaje (1988).
The finding of 6.3% for Klebsiella spp in this study is similar to the 8% prevalence which were found from a study done in Uganda (Yoshimine., et al. 2001) but higher than the 1% found by Ozyilmaz., et al. (2005). E. coli and S. marcescens accounted for 6.3% of the total isolated bacteria which are higher in studies done by Buenviaje (1988) who found 3.6% prevalence and Nagalingam., et al. (2005) who found 1% prevalence of E. coli from sputum samples.
The 1.6% prevalence of P. aeruginosa and Proteus mirabilis is comparable to the 3.6% prevalence of Proteus spp found by Buenviaje (1988) and the 4% prevalence of P. aeruginosa found by Nagalingam., et al. (2005). The 7.9% prevalence of Acinetobacter spp. is higher than when it is compared to the 3.6% prevalence found by Buenviaje (1988).
In the present study, S. pneumoniae was found to be the most frequent pathogen as the etiology of childhood pneumonia. Un-expected finding was that S. pyogenes was accounted for 14.3%. But, S. pyogenes is considered as a less common agent of community-acquired pneumonia (Bartlett and Mundy, 1995). It is noteworthy that E. cloacae was the most frequently isolated Gram-negative enteric bacteria.
There were more Gram-negative bacteria than Gram-positive bacteria isolated from specimens of patients with previous antibiotic treatment than those patients without previous antibiotic treatment which might indicate sampling prior to antibiotic treatment can influence the recovery of pathogens.
Antibiotic resistance among pneumococci is increasing and is of concern because pneumococcus is an important cause of severe community-acquired pneumonia in children and because penicillin and macrolide resistance are increasingly linked (BTS, 2002). Antibiotic resistance varies with time, patient age and geographic area. In the present study, the Streptococcus pneumoniae isolates were 100% susceptible to penicillin and ampicillin.
This finding is comparable to the study done in Jimma by Gebre-Selassie (2002) who found 7.7% resistance to penicillin and ampicillin. But, this finding is very low when compared to the spreading and alarming resistance observed in countries like Tunisia, Algeria, Uganda and Korea where reports of the early 2000s showed 53.7%, 46.2%, 83.5% and 79.7%, respectively (Ramdani-Bouguessa and Rahal, 2003; ANSORP, 1999).
Studies in South Asian countries showed that the prevalence of Penicillin non-susceptible Streptococcus pneumoniae from invasive infections in children was 1.3% in Algeria (Ramdani-Bouguessa and Rahal, 2003). All isolates of S. pneumoniae from lower respiratory tract in Trinidad (Orrett, 2005) showed no resistance to penicillin and ampicillin which is similar to the finding of this study. Resistance of S. pneumoniae to other drugs found in this study was as follows: 11.1% for gentamycin and clindamycin, 22.2% for norfloxacin, and 33.3% for SXT and tetracycline.
Prevalence of resistance was reported in Algeria as follows: 26.4% for tetracycline, 25.7% for Co-trimoxazole, and 4.6% for chloramphenicol (Ramdani-Bouguessa and Rahal, 2003). Resistance to tetracycline, Co-trimoxazole, chloramphenicol was reported to be 25%, 38.3% and 36.7%, respectively in a study done in Trinidad (Orrett, 2005). No resistance to beta-lactams (penicillin, cefotaxime, ampicillin and amoxicillin), erythromycin, chloramphenicol and vancomycin was observed in this study.
In the present study, all isolates of S. pyogenes were sensitive to amoxicillin, clindamycin, ciprofloxacin, cefotaxime, and chloramphenicol. Relatively, resistance is detected for ampicillin, erythromycin, penicillin, vancomycin, SXT, tetracycline and gentamycin. The S. aureus isolates, 7/8 (87.5%) and 6/8 (75.0%) were resistant to penicillin and ampicillin, respectively. This finding corresponds to the study done in Addis Ababa by Geyid., et al. (1991) who found 83.1% of S. aureus strains resistant to penicillin G. No resistance to ciprofloxacin, norfloxacin and vancomycin was observed in this study. No resistance of S. aureus to vancomycin and clindamycin was reported by Geyid., et al. (1991) and Gebre-Selassie (2002). But, in the present study, 12.5% isolates of S. aureus were found to be resistant to clindamycin.
Clinical experience suggested that development of resistance to vancomycin by S. aureus was difficult, despite occasional reports of resistance at low-level. An in vitro demonstration in 1992 showed that the resistance genes from Enterococcus could be passed to S. aureus and subsequently expressed, thus conferring vancomycin resistance. This was a matter of concern, but until 2002 such a transfer had not been reported in wild strains. In 2002, a newly reported Vancomycin-resistant Staphylococcus aureus was isolated from the catheter tip of a renal dialysis patient in Michigan (Chang., et al. 2003).
The resistance patterns of S. aureus to tetracycline, erythromycin and gentamycin were found to be 62.5%, 12.5%, and 25.0%, respectively. The finding in the present study showed similarities to a report by Megerssa (1993) which was 11.1% for gentamycin but with marked difference with the report of Gebre-Selassie (2002) which was 0%. The increased resistance of S. aureus to penicillin (87.5%) and ampicillin (75.5%) in this study showed that there is an increase in drug resistant strains similar to an increase in many areas in the country and even in the world.
There is some evidence that increased use of third-generation cephalosporins leads to resistance in Gram-negative enteric bacteria, especially Citrobacter, Enterobacter, Serratia and Providencia species, and also to emergence of extended-spectrum beta-lactamase (ESBL)-prodcing bacteria (Jukemura., et al. 2007). The finding that Gram-negative enteric bacteria showed 100% resistance to penicillin and clindamycin, 97% resitance to ampicillin and 87% resitance to erythromycin was not unexpected, since these drugs target Gram-positive cocci.
The susceptibility of Gram-positive bacteria to ciprofloxacin and chloramphenicol and Gram-negative microbes to norfloxacin and ciprofloxacin indicates that these bacteria have not yet developed resistance to these drugs. Multi-drug resistance (i.e. resistance to 2 or more drugs) was observed in most of the isolated bacteria. S. aureus showed resistance in 10 of the 13 drugs tested, ranging from 87.5% to 12.5%; S. pneumoniae in 5 of the 13 antibiotics tested; S. pyogenes in 8 of 11 and almost all isolated Gram-negative bacteria showed multiple drug resistance.
Conclusion
S. pneumoniae was found as the most prevalent pathogen of childhood CAP but Penicillin-resistant S. pneumoniae was considerably low. Multi-drug resistance (i.e. resistance to 2 or more drugs) was observed in most of the isolated bacteria. S. aureus showed resistance in 10 of the 13 drugs tested, ranging from 87.5% to 12.5%; S. pneumoniae in 5 of the 13 antibiotics tested; S. pyogenes in 8 of 11 and almost all isolated Gram-negative bacteria showed multiple drug resistance.
Acknowledgments
My sincere thanks to staff members in Bacteriology Laboratory Unit of Tikur Anbassa Hospital for their assistance in technical aspects during laboratory work. I am also indebted to Nurses working in Pediatric Out-Patient Department Emergency Unit for their collaboration in collecting sputum samples from children.
My sincere thanks to staff members in Bacteriology Laboratory Unit of Tikur Anbassa Hospital for their assistance in technical aspects during laboratory work. I am also indebted to Nurses working in Pediatric Out-Patient Department Emergency Unit for their collaboration in collecting sputum samples from children.
My gratitude also goes to the Department of Microbiology, Immunology and Parasitology for providing me API test kits and the unreserved support in facilitating good working environment and to Addis Ababa University Research and Graduate Programs Office for its financial support to conduct the study.
References
- Aderaye G. “The value of sputum Gram stains in the diagnosis of pneumococcal pneumonia”. Ethiopian Medical Journal 32.3 (1994): 167-171.
- Song JH., et al. “Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study”. Clinical Infectious Diseases 28.6 (1999): 1206-1211.
- Bartlett JG and Mundy LM. “Current Concepts: Community-Acquired Pneumonia”. The New England Journal of Medicine 333 (1995): 1618-1624.
- Bauer AW., et al. “Antibiotic susceptibility testing by a standardized single disk diffusion method”. American Journal of Clinical Pathology 45.4 (1966): 493-496.
- Bergey’s Manual of Systematic Bacteriology. (2010). The Firmicutes. Second Edition, Volume 3 Springer Dordrecht Heidelberg London New York.
- Black AD. “Community-acquired pneumonia - a clinical approach to assessment and management”. South African Family Practice 50.3 (2008): 15-23.
- Bochud PY., et al. “Community-acquired pneumonia. A prospective outpatient study”. Medicine 80.2 (2001): 75-87.
- BTS. “Guidelines for the management of community-acquired pneumonia in childhood: British thoracic society standards of care committee”. Thorax 57 (2002): il-i24.
- Buenviaje MB. “Quantitative sputum culture and Gram stain: Pulmonary infection vs. Colonization”. Journal of Microbiology and Infectious Diseases 18 (1988): 28-35.
- Fang GD., et al. “New and emerging etiologies for community-acquired pneumonia with implications for therapy: A prospective multicenter study of 359 cases”. Medicine 69.5 (1990): 307-317.
- Garcia-Vazquez E., et al. “Assessment of the Usefulness of Sputum Culture for Diagnosis of Community-Acquired Pneumonia Using the PORT Predictive Scoring System”. Archives of Internal Medicine 164 .16 (2004): 1807-1811.
- Gebre-Selassie S. “Patterns of isolation of common Gram positive bacterial pathogens and their susceptibilities to antimicrobial agents in Jimma hospital”. Ethiopian Medical Journal 40.2 (2002): 115-127.
- Geyid A and Lemeneh Y. “The incidence of methicilin resistant S. aureus strains in clinical specimens in relation to other beta-lactamase producing and multiple drug resistance properties in Addis Ababa”. Ethiopian Medical Journal 29.4 (1991): 149-161.
- Guclu AU., et al. “Polymerase chain reaction vs. conventional culture in detection of bacterial pneumonia agents”. Annals of Microbiology 55 (2005): 313-316.
- Jukemura EM., et al. “Control of Multi-resistant bacteria and ventilator-associated pneumonia: Is it possible with changes in antibiotics?” Brazilian Journal of Infectious Diseases 11.4 (2007): 418-422.
- Lahti E., et al. “Induced sputum in the diagnosis of childhood community-acquired pneumonia”. Thorax 64.3 (2009): 252-257.
- Lutfiyya MN., et al. “Diagnosis and treatment of community-acquired pneumonia”. American Family Physician 73 (2006): 442-450.
- March., et al. “Signs and Symptoms Indicative of Community-Acquired Pneumonia in Infants under Six Months”. Brazilian Journal of Infectious Diseases 9.2 (2005): 150-155.
- Marrie TJ. “Pneumococcal pneumonia: epidemiology and clinical features”. Seminars in Respiratory Infections 14.3 (1999): 227-236.
- McIntosh K. Community–Acquired Pneumonia in Children. The New England Journal of Medicine 346 (2002): 429-437.
- Megerssa D. “Drug resistance: A retrospective survey in Illubabor region”. Bull of Jimma Institute of Health Science 3 (1993): 51-60.
- Michelow IC., et al. “Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children”. Pediatrics 113.14 (2004): 701-707.
- Nagalingam NA., et al. “A Cross-sectional Study of Isolates from Sputum Samples from Bacterial Pneumonia Patients in Trinidad”. Brazilian Journal of Infectious Diseases 9.3 (2005): 231-240.
- Nagendra S. “Sampling Variability in the Microbiological Evaluation of Expectorated Sputa and Endotracheal Aspirates”. Journal of Clinical Microbiology 39.6 (2001): 2344-2347.
- Nascimento-Carvalho CMC. “Etiology of Childhood Community-Acquired Pneumonia and Its Implications for Vaccination”. Brazilian Journal of Infectious Diseases 5 (2001): 87-97.
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing 11th information supplement Mloo-511 NCCLS, Wayne, Pa. (2001):
- Orrett FA. “Pneumococcal Infections in Trinidad: Patterns of antimicrobial susceptibility: 1994-2002”. Japanese Journal of Infectious Diseases 58 (2005): 20-24.
- Ozyilmaz E., et al. “Major Bacteria of Community-Acquired Respiratory Tract Infections in Turkey”. Japanese Journal of Infectious Diseases 58.1(2005): 50-52.
- Ramdani-Bouguessa N and Rahal K. “Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolated in Algiers, Algeria”. Antimicrobial Agents and Chemotherapy 47.2 (2003): 824-826.
- Roson B., et al. “Prospective Study of the Usefulness of Sputum Gram Stain in the Initial Approach to Community-Acquired Pneumonia Requiring Hospitalization”. Clinical Infectious Diseases 31.4 (2000): 869-874.
- Shah AA., et al. “Study on the prevalence of Enterobacteriaceae in hospital- acquired and community-acquired infections”. Pakistan Journal of Medical Research 41 (2002):
- UNICEF. Pneumonia: The forgotten killer of children. (2006):
- World Health Organization. “Program for the control of Acute Respiratory Infections, 6th programme report ARI 94.33”. WHO Geneva (1994):
- Yamazaki T., et al. “Epidemiology of Community-Acquired Pneumonia in Children”. Pediatrics 115 (2005): 517.
- Yoshimine H., et al. “Community- Acquired Pneumonia in Ugandan Adults: Short-term Parenteral Ampicillin Therapy for Bacterial Pneumonia”. The American Journal of Tropical Medicine and Hygiene 64.3 (2001): 172-177.
Citation:
Befekadu Teshome and Solomon Gebresselassie. “Bacterial Etiologies of Community-Acquired Pneumonia in Children and Their Antimicrobial Susceptibility Patterns”. Clinical Biotechnology and Microbiology 1.4 (2017): 172-185.
Copyright: © 2017 Befekadu Teshome and Solomon Gebresselassie. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.






























 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.