Research Article
Volume 1 Issue 6 - 2018
Isolation and Identification of a New Bacterial Strains Degrading Paracetamol Isolated from Yemeni Environment
1Microbiology Division, Biology Department, Faculty of Science, Sana'a University, Yemen
2Medicinal Chemistry Department, Faculty of Pharmacy, Sana'a University, Yemen
3Chemistry Department, Faculty of Science, Sana'a University, Yemen
2Medicinal Chemistry Department, Faculty of Pharmacy, Sana'a University, Yemen
3Chemistry Department, Faculty of Science, Sana'a University, Yemen
*Corresponding Author: Wadhah H Edrees, Microbiology Division, Biology Department, Faculty of Science, Sana'a University, Yemen.
Received: December 24, 2017; Published: January 09, 2018
Abstract
Paracetamol has emerged as an important environment pollutants due to its wide usage. This study carried out to isolate and identify of a novel paracetamol-degrading bacteria from the contaminated site of pharmaceutical wastewater at Sana'a City, Yemen. The enrichment media and selective media were used to isolate and purify the bacteria from wastewater samples. The isolated bacteria were identified according to phenotypic and genotypic characterization using 16S rRNA sequence analysis. The results showed that the three bacterial strains and signed as an STB1, STB3 and STB5.
The STB1 strain was identified as Pseudomonas aeruginosa strain DSM 50071 with similarity 92%, while the STB3 and STB5 were identified as P. aeruginosa strain NBRC 12689 with similarity 89% and 99%, respectively. The optimal conditions for paracetmol degradation by all stains were recorded at 30˚C and pH 7.0. Also, the paracetamol degradation was found to be influenced by an increase in the bacterial cell concentration. The STB1, STB3, and STB5 strains were degraded 68.6%, 51.3% and 79.4% of 4000 mg/L paracetamol, respectively, in 120h. The results performed that the degradation potential of the three isolated bacterial strains possibly played a major active role in the paracetamol removal processes.
Keywords: Bacteria Isolation;Biodegradation; Paracetamol; Physiological characterization
Introduction
Paracetamol or acetaminophen is a common analgesic and an anti-inflammatory commonly used as a non-prescription drug and sold as over-the-counter drug worldwide [1]. The paracetamol consumption has increased throughout the world in the last decades. In England, it was consumed as one of the top three drugs prescribed, while in the USA, it was consumed as one of the top 200 prescriptions [2,3]. It was ranked as the second prescription drugs throughout the year 2008 in Kuwait [4]. In Yemen, it was also ranked as first of top ten drugs produced by local industries and one of the top ten drugs imported [5].
Paracetamol is one of the most often detected drug products in the aquatic environment. It was 0.298 μg/L reported in drinking water [6], 6.5 μg/L in groundwater [7], 15.7 µg/L in surface water [8], 0.246 mg/L in sewage water [9], and 1.367 mg/L in wastewater [10]. Therefore, the public awareness about its potential risk and effect of paracetamol on the environment and human health has raised [11].
Previous studies on removal of paracetamol from wastewater mainly focused on chemical methods including oxidation processes [12]. The cost of the chemical agent, the energy source, and the generation of secondary pollutants due to the use of excessive chemicals are the major drawbacks to using these methods on removing the paracetamol from wastewaters [13]. So, the biodegradation of paracetamol compound is being considered as an environmentally friendly and low-cost option and has been demonstrated to have the potential to remove this substance by degrading it to nontoxic compounds [14].
This study describes the isolation, identification, and characterization of a new paracetamol degrading bacteria which isolated from wastewater contaminated site of a Yemen Drug Company for Industrial and Commercial (YEDCO) situated at Sana'a City, Yemen. The wastewater was generated by the factory containing a high concentration of paracetamol through the manufacturing processes.
Materials and Methods
Study Design
This study was conducted at YEDCO factory located at Sana'a City, Yemen. 50 samples were collected from different places in and around this factory such as production units wastewater, wastewater channels, and wastewater influent to the sewer. The experimental study carried out at the quality control department at YEDCO for chemical and microbiological analysis during 10 months, from period November 2016 to August 2017.
This study was conducted at YEDCO factory located at Sana'a City, Yemen. 50 samples were collected from different places in and around this factory such as production units wastewater, wastewater channels, and wastewater influent to the sewer. The experimental study carried out at the quality control department at YEDCO for chemical and microbiological analysis during 10 months, from period November 2016 to August 2017.
Chemicals and Cultivation Medium
The standard of paracetamol was purchased from Anqiu Lu'an Pharmaceutical Co. Ltd. (China) and all other chemicals were of the highest purity commercially available. The applied mineral salts medium (MSM) contained the following components (per liter): 0.5g of K2HPO4, 0.5g of KH2PO4, 0.01g of NaCl, 0.2g of MgCl2·6H2O, 0.02g of CaCl2, 0.339 mg of MnSO4, 0.428 mg of ZnSO4, 0.347 mg of (NH4)6Mo7O24·4H2O, 0.4 mg of CoCl2·6H2O, 10 mg of EDTA. The pH was adjusted to 7.2–7.4 with 8M NaOH, the solution and autoclaved at 121˚C for 15 min. Paracetamol was sterilized by filtration paper (0.22 µm) and added to the medium as the sole energy and carbon sources in different concentration [3].
The standard of paracetamol was purchased from Anqiu Lu'an Pharmaceutical Co. Ltd. (China) and all other chemicals were of the highest purity commercially available. The applied mineral salts medium (MSM) contained the following components (per liter): 0.5g of K2HPO4, 0.5g of KH2PO4, 0.01g of NaCl, 0.2g of MgCl2·6H2O, 0.02g of CaCl2, 0.339 mg of MnSO4, 0.428 mg of ZnSO4, 0.347 mg of (NH4)6Mo7O24·4H2O, 0.4 mg of CoCl2·6H2O, 10 mg of EDTA. The pH was adjusted to 7.2–7.4 with 8M NaOH, the solution and autoclaved at 121˚C for 15 min. Paracetamol was sterilized by filtration paper (0.22 µm) and added to the medium as the sole energy and carbon sources in different concentration [3].
Isolation and Purification of Bacteria and Culture Conditions
One mL of each sample was transferred separately into bottle contained 90 mL of trypticase soy broth (TSB) and incubated at 30˚C for 3 days. After the incubation period, ten mL of TSB media was inoculated into a bottle containing 90 mL of MSM with different concentration of paracetamol (500–2000 mg/L) and incubated at 30˚C for 5 days [15,16].
One mL of each sample was transferred separately into bottle contained 90 mL of trypticase soy broth (TSB) and incubated at 30˚C for 3 days. After the incubation period, ten mL of TSB media was inoculated into a bottle containing 90 mL of MSM with different concentration of paracetamol (500–2000 mg/L) and incubated at 30˚C for 5 days [15,16].
Then, 1 mL of MSM was transferred to MSM agar containing 500 mg/L of paracetamol and incubated 5 days at 30˚C. The colonies showing growth on the plate were purified on trypticase soy (TSA) and after several transfers on MSM agar containing paracetamol and checked again on TSA to be pure. Pure colonies isolated were tested for growth on higher concentrations of paracetamol (500–3000 mg/L) in MSM agar. The isolated strains showing best growth of the compounds under study were selected and characterized further [15].
Identification of Isolated Bacteria
The isolated bacteria were identified according to its phenotypic and genotypic characteristics. Preliminary characterizations were based on morphological, physiological and biochemical tests [17].Further identification was completed by 16S rDNA sequencing.Genomic DNAs of the harvested cells were extracted using a DNeasy® Blood & Tissue kit (QIAGEN, Germany), and the 16S rRNA was amplified by means of PCR (Eppendrof, Germany) using the bacteria universal primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (I) (5′-GGTTACCTTGTTACGACTT-3′).
The isolated bacteria were identified according to its phenotypic and genotypic characteristics. Preliminary characterizations were based on morphological, physiological and biochemical tests [17].Further identification was completed by 16S rDNA sequencing.Genomic DNAs of the harvested cells were extracted using a DNeasy® Blood & Tissue kit (QIAGEN, Germany), and the 16S rRNA was amplified by means of PCR (Eppendrof, Germany) using the bacteria universal primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (I) (5′-GGTTACCTTGTTACGACTT-3′).
PCR mixture contained 5 µL of 5X Q5 reaction buffer, 0.2 µL of 10 mM dNTPs, 100 pM of forward primer and 100 pM of reverse primer, 5 ng/uL of DNA template, 0.01 µL/µL of Q5 high-fidelity DNA polymerase (NEB, M041S) and the volume completed up to 25 µL by nuclease-free water [18].
Amplification was started with an initial denaturation at 98˚C for 5 min, followed by 35 cycles at 98˚C for 20 s, 58˚C for 25 s, and 72˚C for 30 s/kb, followed by 8 min at 72˚C for a final extension. The resulting 16s rRNA gene sequences were compared to database NCBI using BLAST program [19]. The multiple alignments of the sequence were performed, and a neighbor-joining phylogenetic tree was constructed using the latest version (2.6.1) of the BLAST program.
Physiological Characterization
The physiological characterizations such as temperature, pH, glucose concentration, and bacteria cell count were investigated as the following:-
The physiological characterizations such as temperature, pH, glucose concentration, and bacteria cell count were investigated as the following:-
Temperature and pH Effect on Paracetamol Biodegradation
The optimum of temperature and pH effects on the biodegradation rate of paracetamol (1500 mg/L) was determined within 72h. For the temperature effect, each bacterium strain was cultivated individually in a volume of 100 mL of MSM media containing paracetamol and incubated at different levels of temperature (15, 20, 25, 30, 35, 40 and 45˚C) for 72h.
The optimum of temperature and pH effects on the biodegradation rate of paracetamol (1500 mg/L) was determined within 72h. For the temperature effect, each bacterium strain was cultivated individually in a volume of 100 mL of MSM media containing paracetamol and incubated at different levels of temperature (15, 20, 25, 30, 35, 40 and 45˚C) for 72h.
However, in the pH effect study, the bacteria strains were cultivated independently in MSM media containing paracetamol and having different levels of pH (4, 5, 6, 6.5, 7, 7.5, 8, 9, and 10) and incubated at 30˚C for 72h. For rate degradation, 5 mL of each culture medium was withdrawn and centrifuged individually at 8000 rpm 30 min. The supernatant was collected in separate clean test tubes and examined for residual paracetamol content [15,16].
Contact Time Effects on Paracetamol Biodegradation
The effect of contact time on paracetamol biodegradation was performed on MSM media for each bacterium strain. Each bacterium isolated was cultured separately on MSM media containing a different concentration of paracetamol (1000‒4000 mg/L) and incubated in different contact time with a label (24, 48, 72, 96, and 120h) at 30˚C. Sampling was done at regular time intervals for residual paracetamol measuring [20].
The effect of contact time on paracetamol biodegradation was performed on MSM media for each bacterium strain. Each bacterium isolated was cultured separately on MSM media containing a different concentration of paracetamol (1000‒4000 mg/L) and incubated in different contact time with a label (24, 48, 72, 96, and 120h) at 30˚C. Sampling was done at regular time intervals for residual paracetamol measuring [20].
Glucose Concentration Effects on Paracetamol Biodegradation
The glucose effecting on paracetamol biodegradation was estimated with different concentration of glucose (0, 1, 3, and 5 mg/L). Each concentration of glucose was added independently to MSM media (100 mL) containing-paracetamol (2500 mg/L) inoculated by each bacterium strain and incubated at 30˚C for 120 h. After each time designated, the sample was analyzed for residual paracetamol [20].
The glucose effecting on paracetamol biodegradation was estimated with different concentration of glucose (0, 1, 3, and 5 mg/L). Each concentration of glucose was added independently to MSM media (100 mL) containing-paracetamol (2500 mg/L) inoculated by each bacterium strain and incubated at 30˚C for 120 h. After each time designated, the sample was analyzed for residual paracetamol [20].
Bacteria Concentration Effects on Paracetamol Biodegradation
The MSM medium containing 2000 mg/L of paracetamol was cultivated by each bacterial strain individually with different initial bacterial concentration (1.0 × 104, 1.0 × 106, and 1.0 × 108 CFU/mL) and incubated at 30˚C for 72h. The residual paracetamol content was assessed for each experimental individually [20].
The MSM medium containing 2000 mg/L of paracetamol was cultivated by each bacterial strain individually with different initial bacterial concentration (1.0 × 104, 1.0 × 106, and 1.0 × 108 CFU/mL) and incubated at 30˚C for 72h. The residual paracetamol content was assessed for each experimental individually [20].
Chemical Analysis Methods
300 mg of working standard of paracetamol was weighed and dissolved in a volumetric flask containing 100 mL of methanol. 5 mL of this solution was transferred to a 100 mL volumetric flask containing 47 mL of methanol and 53 mL of distilled water and mixed well.Then, the peraperd solution was filtered by 0.22 μm nylon membrane filter before used [21].
300 mg of working standard of paracetamol was weighed and dissolved in a volumetric flask containing 100 mL of methanol. 5 mL of this solution was transferred to a 100 mL volumetric flask containing 47 mL of methanol and 53 mL of distilled water and mixed well.Then, the peraperd solution was filtered by 0.22 μm nylon membrane filter before used [21].
Sample Analyzed
The residual concentration of paracetamol was assessed by using HPLC (PerkinElmer, USA). The mobile phase with a flow rate of 1.0 mL/min was composed of acetonitrile: water (47: 53 v/v). The separation was performed at 30˚C by RP–8 column with dimension (5 μm, 4.6 × 250 mm). The injection volume was 20 μm and detection wavelength was fixed at 275 nm [21].
The residual concentration of paracetamol was assessed by using HPLC (PerkinElmer, USA). The mobile phase with a flow rate of 1.0 mL/min was composed of acetonitrile: water (47: 53 v/v). The separation was performed at 30˚C by RP–8 column with dimension (5 μm, 4.6 × 250 mm). The injection volume was 20 μm and detection wavelength was fixed at 275 nm [21].
Statistical Analysis
The obtained data were performed by statistical analysis using IBM SPSS Statistics software (version 18.0, 2009). The statistical analysis of variance between biodegradation efficiency and isolated bacterial strains were compared by Kruskal-Wallis test. Also, the analysis of variance between biodegradation efficiency and physiological characterizations (glucose and microbial concentration) were compared by Friedman test. However, the correlation coefficient between degradation and physiological characterizations (pH and temperature) were performed by using Pearson test. Values (P < 0.01) was considered statistically significant.
The obtained data were performed by statistical analysis using IBM SPSS Statistics software (version 18.0, 2009). The statistical analysis of variance between biodegradation efficiency and isolated bacterial strains were compared by Kruskal-Wallis test. Also, the analysis of variance between biodegradation efficiency and physiological characterizations (glucose and microbial concentration) were compared by Friedman test. However, the correlation coefficient between degradation and physiological characterizations (pH and temperature) were performed by using Pearson test. Values (P < 0.01) was considered statistically significant.
Results
Three bacteria strain named STB1, STB3, and STB5 that capable of degrading paracetamol were isolated from the pharmaceutical wastewater samples. These bacteria were gram-negative, slimy colonies, and non-spore forming. All strains were motile rods and positive for the catalase, oxidase, and nitrate reduction test. While, the citrate utilization, Vogues Proskauer, indole production, H2S production, urease, DNase, and methyl-red test were negative.
From the current results, Figure 1 showed the color change on the MSM broth containing 4000 mg/L of paracetamol by isolated bacteria strains. The change in media color indicated that there were new transformation products formed during the degradation.
The 16S rRNA was amplified from all samples specifically by the selected primers and the bands show the expected size (approximately 1.4 kb) according to the reference marker (Figure 2).
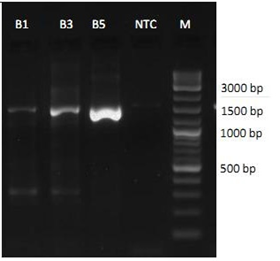
Figure 2: 1.7% agarose gel shows the resolved bands of the 16S rRNA from the bacterial strains; NTC = No Template Control.
The results of 16S rRNA sequence alignment and phylogenetic tree analysis revealed that the 16S rRNA sequence of strains of STB1, STB3, and STB5 belonged to the genus P. aeruginosa. An 1174 base per (bp) 16S rRNA gene sequence that was obtained from strain STB1 and analyzed shows 92% similarity to the 16S rRNA gene sequence of P. aeruginosa strain DSM 50071 (Figure 3).
Moreover, a 1223 bp 16S rRNA gene sequence that was obtained from strain STB3 and analyzed shows 89% similarity to the 16S rRNA gene sequence of P. aeruginosa strain NBRC 12689 (Figure 4). A 749 bp 16S rRNA gene sequence that was obtained from strain STB5 and analyzed shows 99% similarity to the 16S rRNA gene sequence of P. aeruginosa strain NBRC 12689 (Figure 5).
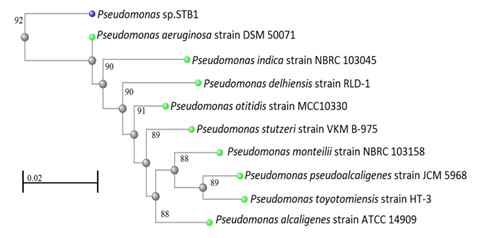
Figure 3: Neighbor-joining phylogenetic tree derived from 16S rRNA gene sequence data showing the positions of Pseudomonas sp. STB1 and related organisms. Bar, 0.02 changes per nucleotide position.
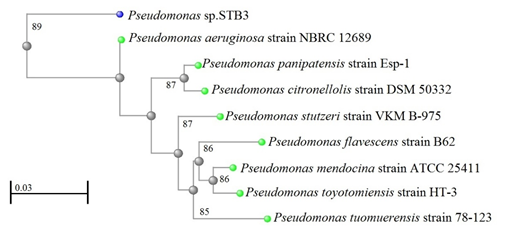
Figure 4: Neighbor-joining phylogenetic tree derived from 16S rRNA gene sequence data showing the positions of Pseudomonas sp. STB3 and related organisms. Bar, 0.03 changes per nucleotide position.
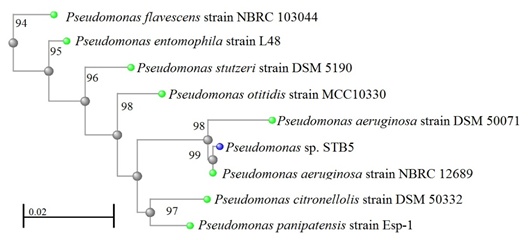
Figure 5: Neighbor-joining phylogenetic tree derived from 16S rRNA gene sequence data showing the positions of Pseudomonas sp. STB5 and related organisms. Bar, 0.02 changes per nucleotide position.
Temperature and pH Effect on the Paracetamol Biodegradation
The results revealed that the optimum temperature was found to be at around 30˚C for paracetamol biodegradation by all strains. The maximum biodegradation of paracetamol was 100% recorded at 30˚C by bacteria strain STB1 and STB5 after 72h. Also, the maximum degradation by STB3 was 93.46%, respectively, reported of 1500 mg/L of paracetamol within 72h (Table 1).
The results revealed that the optimum temperature was found to be at around 30˚C for paracetamol biodegradation by all strains. The maximum biodegradation of paracetamol was 100% recorded at 30˚C by bacteria strain STB1 and STB5 after 72h. Also, the maximum degradation by STB3 was 93.46%, respectively, reported of 1500 mg/L of paracetamol within 72h (Table 1).
However, it can still be considered to show relatively high degradation efficiency in a wide range of temperature (25–35˚C). Whereas, the decreasing effects on paracetamol degradation was recorded at temperatures greater than 40˚C or smaller than 20˚C for all strains (Table 1).
| Temperature (˚C) | The paracetamol reduction (%) after 72h | P value | ||
| STB1 | STB3 | STB5 | ||
| 15 | 27.93 | 21 | 31.42 | < 0.01* |
| 20 | 49.2 | 40.57 | 53 | |
| 25 | 78.96 | 72.5 | 79.16 | |
| 30 | 100 | 93.46 | 100 | |
| 35 | 84.4 | 77.64 | 85.3 | |
| 40 | 47.25 | 42.81 | 51 | |
| 45 | 27.91 | 20.49 | 30.1 | |
Table 1: The effect of different temperatures on paracetamol biodegradation by isolated bacterial strains.
*. Correlation is significant at the 0.01 level (2-tailed).
*. Correlation is significant at the 0.01 level (2-tailed).
Moreover, the results in Table 2 showed that the optimum pH for paracetamol degradation was observed at pH 7.0 for all isolated bacterial strains. Also, it was reported that the complete degradation of 1500 mg/L of paracetamol was recorded by STB1 and STB5 after 72h. In addition, 93.46% of paracetamol was degraded by STB3, respectively, at the same pH and time as listed in Table 2. The degradation rate of paracetamol decreased in an experimental study at pH greater than 8.0 or smaller than 6.0. However, it can still be considered to show relatively high degradation efficiency in a wide range of pH (6.0–8.0) and performed the best in the pH range of 6.5–7.5 for all strains (Table 2).
| pH | The paracetamol reduction (%) after 72h | P value | ||
| STB1 | STB3 | STB5 | ||
| 4 | 19 | 16 | 20 | < 0.01* |
| 5 | 42 | 38.14 | 43 | |
| 6 | 63.54 | 58 | 62 | |
| 6.5 | 97 | 89.53 | 96.86 | |
| 7 | 100 | 93.46 | 100 | |
| 7.5 | 96.31 | 88.79 | 95.92 | |
| 8 | 85.17 | 80.15 | 86.14 | |
| 9 | 37 | 32.7 | 38 | |
| 10 | 15 | 10 | 16 | |
Table 2: The effect of different levels of pH on paracetamol biodegradation by isolated bacterial strains.
*. Correlation is significant at the 0.01 level (2-tailed).
*. Correlation is significant at the 0.01 level (2-tailed).
Effect of Contact Time on Biodegradation of Different Concentration of Paracetamol
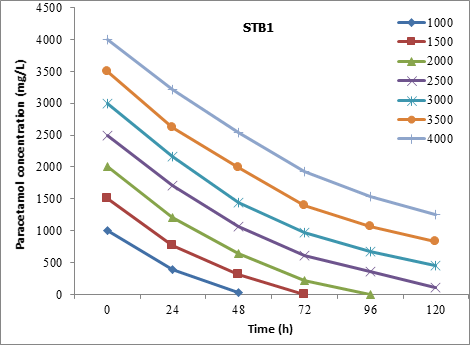
The STB1 strain has completely degraded the paracetamol at a concentration of 1500 mg/L within 72h. Also, it was recorded that the 84.73% and 68.6% of 3000 mg/L and 4000 mg/L, respectively, of paracetamol were degraded at 120h (Figure 6).
The STB1 strain has completely degraded the paracetamol at a concentration of 1500 mg/L within 72h. Also, it was recorded that the 84.73% and 68.6% of 3000 mg/L and 4000 mg/L, respectively, of paracetamol were degraded at 120h (Figure 6).
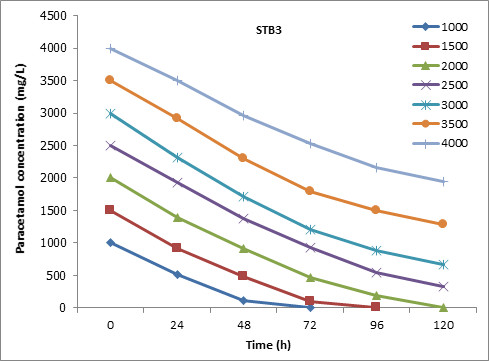
Furthermore ,the strain STB3 was reported to degrade 93.46% of 1500 mg/L paracetamol after 72h of incubation. At high concentration of paracetamol, the STB3 was recorded to degrade 86.92%, 63.22%, and 51.3% of 2500, 3000, and 4000 mg/L, respectively, of paracetamol within 120h of incubation (Figure 7).
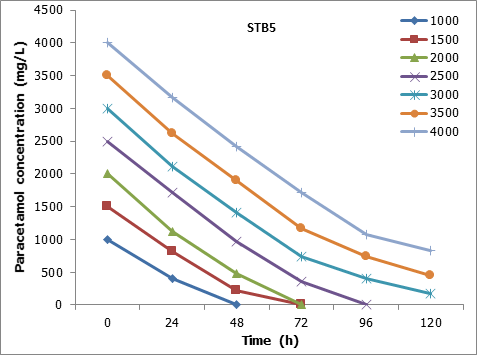
The results showed that the STB5 was 100% degraded the 2000 mg/L of paracetamol at 72h. Also, 94.13% and 79,4% of 3000 and 4000 mg/L, respectively, of paracetamol were degraded at 120h (Figure 8).
The Kruskal-Wallis test showed that there is not a significant difference (P = > 0.01) in the biodegradtion effeciency between the STB1, STB3, and STB5 starins.
Effect of Glucose on Paracetamol Biodegradation
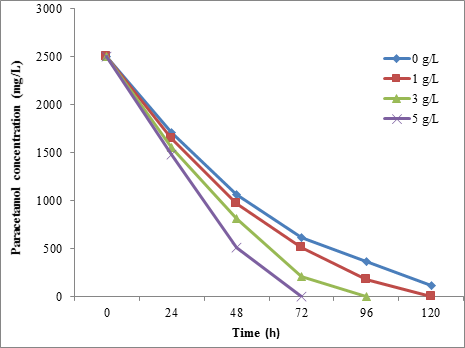
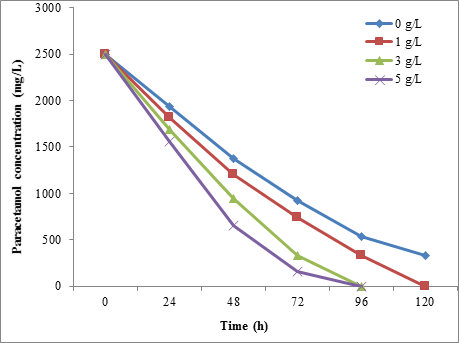
From the present results, the glucose was found to be accelerated the paracetamol biodegradation by all isolated bacterial strains. It was found that the STB1 was degraded 100% of 2500 mg/L of paracetamol when added 5 g/L of glucose to the media. Without glucose, the STB1 was degraded 75.40% of 2500 mg/L of paracetamol within 72h (Figure 9).
From the present results, the glucose was found to be accelerated the paracetamol biodegradation by all isolated bacterial strains. It was found that the STB1 was degraded 100% of 2500 mg/L of paracetamol when added 5 g/L of glucose to the media. Without glucose, the STB1 was degraded 75.40% of 2500 mg/L of paracetamol within 72h (Figure 9).
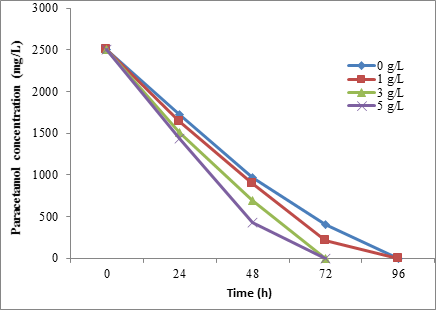
However, the results showed that the paracetamol biodegradation was increased with increasing the glucose concentration for BTS3. At 5 mg/L of glucose, 93.92% and 100% of 2500 mg/L paracetamol were degraded by BTS3 and STB5 strains, respectively, within 72h (Figure 10 and Figure 11). Also, the Friedman test showed a significant difference (P = < 0.01) between the glucose concentration and biodegradation effecincy of all tested strains.
Effect of Bacterial Cells Concentration on Paracetamol Biodegradation
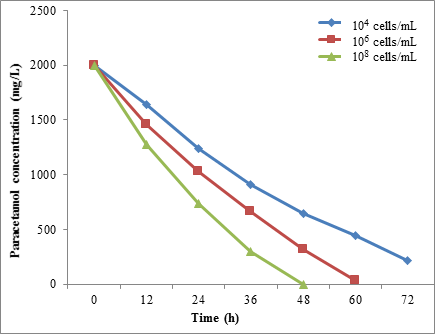
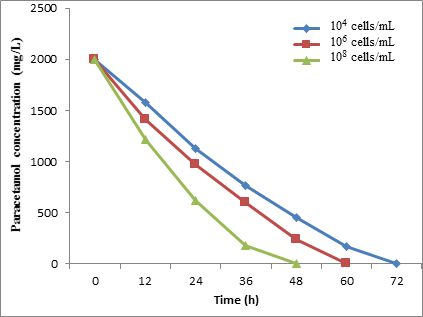
The results showed a different rate of paracetamol degradation with a different concentration of microbial cells. When the microbial concentration cells of STB1 were increased from 1.0 × 104 to 1.0 × 108 cells/mL, the paracetamol degradation was increased from 67.95% to 100%, respectively, after 48 h of incubation (Figure 12).
The results showed a different rate of paracetamol degradation with a different concentration of microbial cells. When the microbial concentration cells of STB1 were increased from 1.0 × 104 to 1.0 × 108 cells/mL, the paracetamol degradation was increased from 67.95% to 100%, respectively, after 48 h of incubation (Figure 12).
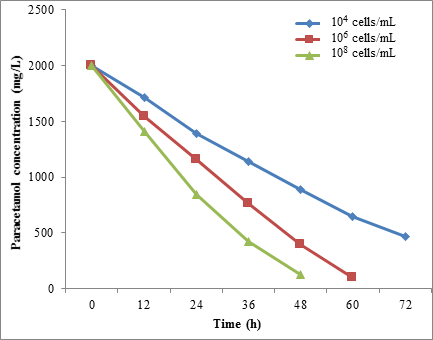
Furthermore, the Figure 13 showed that the STB3 degraded 67.85% of paracetamol at 1.0 × 104 cells/mL within 60h. In addition, the complete degradation occurred when microbial cells increased to 1.0 × 108 cells/mL. Also, the complete (100%) degradation of paracetamol occurred after 60 and 48h for the cell concentration 1.0 × 108 cells/mL STB5 (Figure 14).
However, the Friedman test showed a significant difference (P = < 0.01) between the bacterial concentration and biodegradation effecincy of all tested strains.
Discussion
Biodegradation is an important role in removing paracetamol from the environment. The results of the present study indicate that the site contaminated with the pharmaceutical effluent containing paracetamol is rich with variety microorganisms which tolerate and use it as the sole source of carbon and energy for growth. Three bacteria strain signed as an STB1, STB3, and STB5 were isolated from this contaminated site. The STB1 strain was identified as Pseudomonas aeruginosa strain DSM 50071 with similarity 92%, while the STB3 and STB5 were identified as P. aeruginosa strain NBRC 12689 with similarity 89% and 99%, respectively.
In similar, several reports were recorded that the Pseudomonas species degrading paracetamol were successfully isolated from a different site contaminated with pharmaceutical wastewater containing paracetamol [15,22,23]. The pH plays a vital role in biodegradation and gives an understanding of degradation process of paracetamol substance. The optimum pH for paracetamol degradation was found at 7.0 for all bacteria stains.
This result coincides with Hu., et al. [22] who revealed that the paracetamol (1200 mg/L) was completely degraded by P. aeruginosa strain HJ1012 at around pH 7.0 on 48h. Also, the optimum pH for the Pseudomonas sp. growth on media was recoded to be between 7.0-8.0 [14, 23]. A reduction in paracetamol biodegradation in more acidic and alkaline media is due to the extreme pH levels that inhibit the bacterial cells growth.
The temperature considers a significant role in microbial growth rate and can influence the degradation of organic compounds through direct effects on enzyme activity [24]. In this study, the effect of temperature was found on bacterial strains and 30˚C was shown to be the most effective temperature for paracetamol biodegradation. This result is in agreement with the findings Hu., et al. [22] who found that the optimum temperature was appeared to be at around 30˚C for paracetamol biodegradation by P. aeruginosa strain HJ1012.
Furthermore, a study by Fang., et al. [14] and Yun., et al. [23] observed that the optimum temperature for the Pseudomonas sp. growth on media was recorded at 30˚C. It was documented that growth and metabolic activity of Pseudomonas sp. Ap-3 was maximum at 30˚C [25]. Some of the experimental studies on the paracetamol biodegradation by Pseudomonas sp. were carried out at a temperature of 30˚C and pH of 7.0 [22].
The present study was detected that the degradation rate of paracetamol was affected by cell concentrations which play a significant role in transport elements. These findings are similar to those of Akay and Tezel, [26] who revealed that the biotransformation of paracetamol was influenced at a different cell concentration of R. erythropolis. Biodegradation of organic substrates provides microorganisms with energy and building materials that are used for growth of new cells, cell maintenance and co-metabolism of other less degradable substances [27].
The result of this study was revalued that the biodegradation on paracetamol (2500 mg/L) in the presence of glucose was increased with increasing glucose amounts. A study by Ahmed., et al. [15] observed that the glucose enhance the Pseudomonas sp. strain ST1 growth in media containing paracetamol (500-5000 mg/L). The presence of glucose provides a source of carbon and energy for the microbes, thus enhancing their activity to utilize the resistant aromatic amines [28].
The result showed that the STB1 and STB5 could perform complete degradation of paracetamol at concentrations of 1500 and 2000 mg/L or below, respectively, within 72h. Whereas the STB3 was reported to degrade 93.46% of 1500 mg/L paracetamol after 72h of incubation. At 4000 mg/L, 68.6%, 51.3% and 79.4% of paracetamol concentration were degraded by STB1, STB3, and STB5, respectively, within 120h of reaction. In a similar study by Ahmed., et al. [15] who reported that the isolated Pseudomonas sp. strain ST1 was able to degrade 76.8% of 4000 mg/L paracetamol within 72h.
Another study by Fang., et al. [14] who observed that the Cupriavidus sp. strain F1, Lysobacter sp. strain F2 and Pseudomonas sp. strain (Figure 2) were able to degrade paracetamol up to 400 mg/L, 2500 mg/L, and 2000 mg/L, respectively. In a similar study by Hu., et al. [22] who reported that the P. aeruginosa strain HJ1012 could completely degrade paracetamol as high as 2200 mg/L within 75 h of reaction.
A study by Zhang., et al. [29] who recorded that the Stenotrophomonas sp. strain (Figure 2) and Pseudomonas sp. strain (F1 and F2) were able to complete degradation of paracetamol at concentrations of 2000, 400, and 2500 mg/L or below in 116h, 60h, and 45h, respectively. Also, it was reported that the Pseudomonas sp. degraded 96% of 1500 mg/L paracetamol in 12h [23]. Pseudomonas species are environmental organisms documented for their ability to degrade aromatic compounds of environmental concern [30]. The adaptability of Pseudomonas species to different organic substrates makes it an attractive organism for its use in biodegradation for wide ranges of organic substances that present in wastewater.
Conclusion
Pharmaceutical wastewater is the most potential sources to isolate microbes that degrading paracetamol. The isolated bacterial strains were showed a high degradation efficiency of paracetamol and could tolerate paracetamol at 4000 mg/L. The rate of paracetamol degradation was also affected by initial pH values, temperatures, and a glucose concentration of culture medium. Therefore, P. aeruginosa may prove to be important in bioremediation and wastewater treatment. A further investigation on paracetamol metabolic pathways involved in biodegradation, identification, and isolation of genes responsible for producing degradative enzymes and sequencing and cloning of such genes may increase further knowledge about the abilities of these bacterial strains towards biodegradation process.
References
- Martindale W. “The complete drug reference, cough suppressants, expectorants, mucolytics and nasal decongestants”. The pharmaceutical Press (2009):
- Sebastine IM and Wakeman RJ. “Consumption and environmental hazards of pharmaceutical substances in the UK”. Process Safety and Environmental Protection81.4 (2003): 229-235.
- Zhang X., et al. “Photo degradation of acetaminophen in TiO2 suspended solution”. Journal of Hazardous Materials 157.2-3 (2008): 300-307.
- Alajmi HM. “Effect of physical, chemical and biological treatment on the removal of five pharmaceuticals from domestic wastewater in laboratory-scale reactors and a full-scale plan”. Ph.D. thesis. University of Newcastle upon Tyne (2014): 50-87.
- Edrees HW., et al. Paracetamol in environment and its degradation by different processes. Lambert Academic Publishing Düsseldorf, Germany (2017):
- Kleywegt S., et al. “Pharmaceuticals, hormones and bisphenol A in untreated source and finished drinking water in Ontario, Canada: Occurrence and treatment efficiency”. Science of the Total Environment 409.8 (2011): 1481-1488.
- Zimmerman MJ. “Occurrence of organic wastewater contaminants, pharmaceuticals, and personal care products in selected water supplies”. Cape Cod, Massachusetts, June 2004: U.S. Geological Survey Open-File Report 1206 (2005): 1-16.
- Lin YA and Tsai T. “Occurrence of pharmaceuticals in Taiwan's surface waters: Impact of waste streams from hospitals and pharmaceutical production facilities”. Science of the Total Environment 407.12 (2009): 3793-3802.
- Gomez MJ., et al. “Pilot survey monitoring pharmaceuticals and related compounds in a sewage treatment plant located on the Mediterranean coast”. Chemosphere 66.6 (2007): 993-1002.
- Thomas KV., et al. “Source to sink tracking of selected human pharmaceuticals from two Oslo city hospitals and a wastewater treatment works”. Journal of Environmental Monitoring 9.12 (2007): 1410-1418.
- Wu S., et al. “Paracetamol in the environment and its degradation by microorganisms”. Applied Microbiology and Biotechnology 96.4 (2012): 875-884.
- Andreozzi R., et al. “Paracetamol oxidation from aqueous solutions by means of ozonation and H2O2/UV system”. Water Research 37.5 (2003): 993–1004.
- Trovo AG., et al. “Photodegradation of the pharmaceuticals amoxicillin, bezafibrate and paracetamol by the photo-Fenton process: Application to sewage treatment plant effluent”. Journal of Photochemistry and Photobiology A: Chemistry 198.2-3 (2008): 215–220.
- Fang W., et al. “Study on bacterial function of high-efficiency paracetamol-degrading aerobic granule. Master's thesis”. Zhejiang University of Technology, Microbiology, China (2011): 1-15.
- Ahmed S., et al. “Isolation and characterization of a Pseudomonas strain that degrades 4-acetamidophenol and 4-aminophenol”. Biodegradation 12.5 (2001): 303–309.
- Gusseme BD., et al. “Degradation of acetaminophen by Delftia tsuruhatensis and Pseudomonas aeruginosa in a membrane bioreactor”. Water Research 45.4 (2011): 1829-1837.
- “Bergey’s Manual of Systematic Bacteriology”. Second edition. Michigan State University; East Lansing, USA (2004):
- Chen L., et al. “Rapid sanger sequencing of the 16S rRNA gene for identification of some common pathogens”. PLoS ONE 9.2 (2014): e88886.
- BLAST program: National Center for Biotechnology Information (NCBI).
- Shourian M., et al. “Efficient phenol degradation by a newly characterized Pseudomonas sp. SA01 isolated from pharmaceutical wastewaters”. Desalination 246.1-3 (2009): 577-594.
- United States Pharmacopoeia 30-NF/25: Monograph: Paracetamol (acetaminophen): The United States Pharmacopeial Convention, Rockville, USA; 2005.
- Hu J., et al. “Degradation of paracetamol by Pseudomonas aeruginosa strain HJ1012”.Journal of Environmental Science and Health, Part A: Toxic/Hazardous Substances and Environmental Engineering 48.7 (2013): 791-799.
- Yun MA., et al. “Isolation and characterization of a acetaminophen degrading bacterium”. Journal of Zhejiang University of Technology 1 (2013): 35-38.
- Pettersson M and Baath E. “Temperature-dependent changes in the soil bacterial community in limed and unlimed soil”. FEMS Microbiology Ecology 45.1 (2003): 13-21.
- Takenaka S., et al. “Complete nucleotide sequence and functional analysis of the gene for 2-aminophenol metabolism from Pseudomonas sp. AP-3”. Archives of Microbiology 174.4 (2000): 265-272.
- Akay C and Tezel U. “Biotransformation of acetaminophen by four phylogenetically distinct bacteria”. Proceedings CRETE 2016, Fifth International Conference on Industrial and Hazardous Waste Management. Chania – Crete – Greece (2016): 27–30.
- Cornelissen G and Sijm DT. “An energy budget model for the biodegradation and co-metabolism of organic substances”. Chemosphere 33.5 (1996): 817-830.
- Khan SA., et al. “Influence of pH, temperature and glucose on biodegradation of 4-aminophenol by a novel bacterial strain, Pseudomonas sp. ST-4”. African Journal of Biotechnology 8.16 (2009): 3827-3831.
- Zhang L., et al. “Degradation of paracetamol by pure bacterial cultures and their microbial consortium”. Applied Microbiology and Biotechnology97.8 (2013): 3687-3698.
- Neumann G., et al. “Simultaneous degradation of atrazine and phenol by Pseudomonas sp. strain ADP: Effect of toxicity and adaptation”. Applied and Environmental Microbiology 70.4 (2004):1907–1912.
Citation:
Wadhah H Edrees., et al. “Isolation and Identification of a New Bacterial Strains Degrading Paracetamol Isolated from Yemeni
Environment”. Clinical Biotechnology and Microbiology 1.6 (2018): 257-270.
Copyright: © 2018 Wadhah H Edrees., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.































 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.