Research Article
Volume 3 Issue 1 - 2018
Prevalence of Multidrug-resistant Salmonella enterica and associated factors among under five children with diarrhea in rural Burkina Faso.
1Laboratoire de Biologie Moléculaire, d’Épidémiologie et de Surveillance des Bactéries et Virus Transmissibles par les Aliments (LaBESTA)/
Centre de Recherche en Sciences Biologiques, Alimentaires et Nutritionnelles (CRSBAN)/Université Ouaga I Pr Joseph KI-ZERBO, 03 BP 7021
Ouagadougou 03, Burkina Faso
2Unité de Formation et de Recherche en Sciences Appliquées et Technologiques (UFR/SAT)/Université de Dédougou, BP 07 Dédougou Burkina Faso
3Centre National de Recherche et de Formation sur le Paludisme (CNRFP), 01 BP 2208 Ouagadougou 01, Burkina Faso
4Institut Des Sciences, 01 BP 1757 Ouagadougou 01, Burkina Faso
2Unité de Formation et de Recherche en Sciences Appliquées et Technologiques (UFR/SAT)/Université de Dédougou, BP 07 Dédougou Burkina Faso
3Centre National de Recherche et de Formation sur le Paludisme (CNRFP), 01 BP 2208 Ouagadougou 01, Burkina Faso
4Institut Des Sciences, 01 BP 1757 Ouagadougou 01, Burkina Faso
*Corresponding Author: René Dembélé, Laboratoire de Biologie Moléculaire, d’Épidémiologie et de Surveillance des Bactéries et
Virus Transmissibles par les Aliments(LaBESTA)/Centre de Recherche en Sciences Biologiques, Alimentaires et Nutritionnelles
(CRSBAN)/Université de Dédougou, BP 176 Dédougou, Burkina Faso.
Receieved: October 23, 2018; Published: November 30, 2018
Abstract
Salmonella enterica is one ofthe major enteric pathogen causing diarrhea among children under 5 years worldwide. In rural settings of Burkina Faso where this infection is strongly associated with the consumption of contaminated food and water, the antimicrobial resistant patterns of Salmonella circulating in these areas are currently not well described. To address this issue, stool samples were used to isolate and identify the pathogen. Antimicrobial susceptibility test was performed to isolate Salmonella. A logistic regression analysis was used to assess the association between different variables and outcome. Odds ratio with 95% CI was computed to determine the presence and the strength of the association.Nine (3.30%) Salmonella sp. were isolated and identified as six different serotypes. Among these strains, 43% were isolated from diarrheal children in association with malnutrition. Strains were identified resistant mainly to aminopenicillins [amoxicillin-clavulanic acid (89%), amoxicillin (100%)] and monobactam [aztreonam (44%)]. High level of resistances to the 3rd generation cephalosporins were noted (ceftriaxone: 56%). Resistances to quinolones (22% resistant to nalidixic acid) and fluoroquinolones (11% resistant to ciprofloxacin) were also reported. Multiple drug resistance (MDR) has been reported for most of the Salmonella serotypes (80%). Restrictions on the irrational use of antibiotics in humans and animals are suggested for reduction of multidrug-resistant Salmonella.
Keywords: Salmonella; Diarrhea; Children; Antimicrobial resistance
abbreviations: MDR: Multiple drug resistance; CI: Confidence Interval; OR: Odds Ratios, WHO: World Health Organization; AMR:
Antimicrobial resistance; EUCAST: European Committee on Antibiotic Susceptibility Testing; ESBL: extended spectrum ß-lactamase; S.:
Salmonella; GI: Gastrointestinal Infections
Introduction
Diarrhea represents a worldwide concern frequently encountered in pediatric medicine. Globally arround 550 million of cases are annually notified. Of these 220 million of cases occur in children under the age of 5 years [1]. According to the World Health Organization (WHO) report, diarrheal illness is also the second leading causes of death in children younger than 5 years. Globally 21% of deaths in children under the age of 5 years results from diarrheal infection [2]. Diarrhea kills more young children than Malaria, AIDS, and Measles combined [3]. In developing countries, Shigella and Salmonella species remain major contributors to acute enteric infection in children. Asia, Africa and latin America had an estimated of 2.5 million deaths each year in children under five years of age [4]. Diarrhea is mainly caused by infection with virus, bacteria or parasite [4]. Bacterial diarrhea is commonly caused by Salmonella enterica, Shigella species, Vibrio cholera, Clostridium difficile, Escherichia coli, Campylobacter jejuni and others. Among these, Salmonella represents one of the zoonotic pathogens, of great importance in public health science as often associated with gastroenteritis [5,6]. From the 2015 World Health Organization (WHO) estimation of the global burden of foodborne diseases, Salmonella ranked first among 22 bacterial, protozoal, and viral agents, reflecting its ubiquitous nature and the severity of illnesses [7].
Even, treatment with antimicrobials is crucial for proper management of severe or invasive human salmonellosis [5], antibiotic therapy is not systematic in case of infantile diarrhea with bacterial etiology [8]. However, when antibiotics are used, the choice of the antibiotic become challenging due to the emergence of resistance to first-line antibiotics (chloramphenicol, trimethoprim–sulfametoxazole, tetracycline, and penicillin A) [8]. Antimicrobial resistance (AMR) expanded within a wide range of infectious agents is a growing public health threat of broad concern to countries and multiple sectors [9]. The resistance to fluoroquinolones, in nontyphoidal Salmonella (NTS) and Shigella species were comparatively lower than in E. coli[9]. However, there were considerable gaps in information on these two bacteria, particularly from areas where they remain of major public health importance [9]. The emergence of multidrug-resistant strains, including isolates resistant to quinolones, have been reported from some African countries [10-12, 5] leading to a major problem associated with the control of diarrhea [13]. Moreover, current information regarding antimicrobial susceptibility pattern of bacteria causing diarrhea in children is limited and thus it is uncertain whether the recommended antibiotics are still effective [14]. Therefore, this study aims to evaluate the prevalence of Multidrug-resistant Salmonella enterica and associated factors in diarrheal children in rural area of Burkina Faso.
Materials and Methods
Study design, period and settings
A prospective cross sectional study was conducted to determine the serotypes and antimicrobial susceptibility of Salmonella species among diarrheic children visiting hospitals in rural settings of Burkina Faso. This study was conducted between July 2009 and June 2010 (during one year) in two remote rural areas, at north (Gourcy, distance 140 km) and western (Boromo, distance 185 km) of the capital Ouagadougou, Burkina Faso (Figure 1). Stool samples were collected from 275 under five years of age; 228 diarrheal children and 47 healthy children (control group).
A prospective cross sectional study was conducted to determine the serotypes and antimicrobial susceptibility of Salmonella species among diarrheic children visiting hospitals in rural settings of Burkina Faso. This study was conducted between July 2009 and June 2010 (during one year) in two remote rural areas, at north (Gourcy, distance 140 km) and western (Boromo, distance 185 km) of the capital Ouagadougou, Burkina Faso (Figure 1). Stool samples were collected from 275 under five years of age; 228 diarrheal children and 47 healthy children (control group).
Specimen collection
Stool samples were taken by trained health staff personnel using a swab transport system (M40 transystemAmies agar gel without charcoal; Copan Italia Spa, Brescia, Italy) and transported to laboratory within 24h of their collection for analysis. Information regarding the age and sex were recorded for each child using a questionnaire.
Stool samples were taken by trained health staff personnel using a swab transport system (M40 transystemAmies agar gel without charcoal; Copan Italia Spa, Brescia, Italy) and transported to laboratory within 24h of their collection for analysis. Information regarding the age and sex were recorded for each child using a questionnaire.
Salmonellaisolation and identification
Selenite broth (Emapol, Pologne) was used for the enrichment of specimens followed by an incubation at 37°C for 18h. Subsequently, samples were cultured on Hecktoen Enteric agar (Liofilchem, Italy) and incubated at 37°C for 24h. The identity of typical-looking Salmonella colonies on Hektoen was examined by using orthonitrophenyl-ß-D-galactopyranoside (ONPG), citrate, mannitol, lysine decarboxylase tests and the Kliger Hajna medium (Liofilchem, Italy). Finally the isolates were confirmed by API 20E (BioMérieux, Marcy l’Etoile, France).
Selenite broth (Emapol, Pologne) was used for the enrichment of specimens followed by an incubation at 37°C for 18h. Subsequently, samples were cultured on Hecktoen Enteric agar (Liofilchem, Italy) and incubated at 37°C for 24h. The identity of typical-looking Salmonella colonies on Hektoen was examined by using orthonitrophenyl-ß-D-galactopyranoside (ONPG), citrate, mannitol, lysine decarboxylase tests and the Kliger Hajna medium (Liofilchem, Italy). Finally the isolates were confirmed by API 20E (BioMérieux, Marcy l’Etoile, France).
Serotyping of Salmonella isolates
All Salmonella isolates were serotyped by the Salmonella Reference Laboratory. Isolates were serotyped with the somatic O and flagellar H anti-sera according to the Kauffman White scheme [15].
All Salmonella isolates were serotyped by the Salmonella Reference Laboratory. Isolates were serotyped with the somatic O and flagellar H anti-sera according to the Kauffman White scheme [15].
Antimicrobial susceptibility testing
All Salmonella strains were subjected to antimicrobial susceptibility testing. It was carried out by disc diffusion method on Müller-Hinton agar (Liofilchem, Italy) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing [16]. Briefly, after depositing the antibiotics, the plates were incubated at +37°C for 18-24h. Nineteen (19) antibiotics belonging to 7 different families were tested : amoxicillin (25 µg), amoxicillin–clavulanic acid (20/10 µg), ceftriaxone (30 µg), cefotaxime (30 µg), cefepime (30 µg), cefixime (10 µg), piperacillin (75 µg), piperacillin–tazobactam (100 +10 µg), imipenem (10 µg), tetracycline (30 µg), chloramphenicol (30 µg), trimethoprim–sulfametoxazole (1.25 ± 23.75 µg), aztreonam (30 µg), colistin sulfate (50 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), gentamycin (15 µg), netilmicin (10 µg), and tobramycin (10 µg) (Bio-Rad, France).
All Salmonella strains were subjected to antimicrobial susceptibility testing. It was carried out by disc diffusion method on Müller-Hinton agar (Liofilchem, Italy) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing [16]. Briefly, after depositing the antibiotics, the plates were incubated at +37°C for 18-24h. Nineteen (19) antibiotics belonging to 7 different families were tested : amoxicillin (25 µg), amoxicillin–clavulanic acid (20/10 µg), ceftriaxone (30 µg), cefotaxime (30 µg), cefepime (30 µg), cefixime (10 µg), piperacillin (75 µg), piperacillin–tazobactam (100 +10 µg), imipenem (10 µg), tetracycline (30 µg), chloramphenicol (30 µg), trimethoprim–sulfametoxazole (1.25 ± 23.75 µg), aztreonam (30 µg), colistin sulfate (50 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), gentamycin (15 µg), netilmicin (10 µg), and tobramycin (10 µg) (Bio-Rad, France).
Antibiotics phenotypes determination
Antibiotyping method involves the simultaneous presence of one or more antibiotic resistance markers. A strain may not wear a resistance marker or wear one or more. When studying the susceptibility of a strain to several antibiotics, its resistance phenotype to antibiotics was determined. If the strain expresses only natural resistances, it is said to belong to the "wild" or sensitive phenotype. If it acquired resistances have changed its sensitivity, it expresses a "resistance phenotype" that can be identified and whose mechanism must be determined. This phenotype is often referred to as initials of antibiotics that have become inactive. A strain is described as multidrug resistant when it is resistant to three antibiotics of different families [17-19].
Antibiotyping method involves the simultaneous presence of one or more antibiotic resistance markers. A strain may not wear a resistance marker or wear one or more. When studying the susceptibility of a strain to several antibiotics, its resistance phenotype to antibiotics was determined. If the strain expresses only natural resistances, it is said to belong to the "wild" or sensitive phenotype. If it acquired resistances have changed its sensitivity, it expresses a "resistance phenotype" that can be identified and whose mechanism must be determined. This phenotype is often referred to as initials of antibiotics that have become inactive. A strain is described as multidrug resistant when it is resistant to three antibiotics of different families [17-19].
Phenotypic detection of ESBL
Strains that were β-lactams resistant were subjected to investigation of extended spectrum ß-lactamase (ESBL) activity according to the recommendations of EUCAST [16]. A disk of amoxicillin-clavulanic acid and two disks of third generation cephalosporins (C3G) (ceftriaxone and cefotaxime) were placed on the bacterial plate separated by a distance of 2 to 3 cm from one another. The presence of ESBL is indicated by a syngergetic effect between the disks, giving rise to an extended halo with the appearance of a “champagne cork” of keyhole.
Strains that were β-lactams resistant were subjected to investigation of extended spectrum ß-lactamase (ESBL) activity according to the recommendations of EUCAST [16]. A disk of amoxicillin-clavulanic acid and two disks of third generation cephalosporins (C3G) (ceftriaxone and cefotaxime) were placed on the bacterial plate separated by a distance of 2 to 3 cm from one another. The presence of ESBL is indicated by a syngergetic effect between the disks, giving rise to an extended halo with the appearance of a “champagne cork” of keyhole.
Data processing
Data were entried and analyzed using the software package Epi Info 7.1.2.0 (Centers for Disease Control and Prevention [CDC], Atlanta). Multivariable logistic regression was used to estimate odds ratios (ORs) with ninety-five percent confidence intervals (95% CI) also calculated. The statistical significance was evaluated using the Fischer exact 2-tailed p value and a p ≤ 0.05 was considered significant.
Data were entried and analyzed using the software package Epi Info 7.1.2.0 (Centers for Disease Control and Prevention [CDC], Atlanta). Multivariable logistic regression was used to estimate odds ratios (ORs) with ninety-five percent confidence intervals (95% CI) also calculated. The statistical significance was evaluated using the Fischer exact 2-tailed p value and a p ≤ 0.05 was considered significant.
Ethical considerations
Permission to conduct the study was obtained from the hospital authorities of Burkina Faso, and informed verbal consent was obtained from the parents/guardians of every child before sample collection. The National Ethical Committee (s) of Burkina Faso (N° 2009-39) approved the study protocol.
Permission to conduct the study was obtained from the hospital authorities of Burkina Faso, and informed verbal consent was obtained from the parents/guardians of every child before sample collection. The National Ethical Committee (s) of Burkina Faso (N° 2009-39) approved the study protocol.
Results and Discussion
Socio‑demographic characteristics
A total of 228 children with diarrhea were included in our study (116 from Boromo Health District and 112 from Gourcy Health District). The age of participants range between 1 and 59 months and mean age was 31 months. From the f all study participants, 39.4% were females while 60.6% were males (Table 1).
A total of 228 children with diarrhea were included in our study (116 from Boromo Health District and 112 from Gourcy Health District). The age of participants range between 1 and 59 months and mean age was 31 months. From the f all study participants, 39.4% were females while 60.6% were males (Table 1).
| Age (months) | Sexe | Number of patients | Total N (%) | |
| Boromo n (%) | Gourcy n (%) | |||
| 0-12 | M | 30 (13.2) | 38 (16.6) | 68 (29.8) |
| F | 28 (12.3) | 22 (9.6) | 50 (21.9) | |
| 13-35 | M | 29 (12.7) | 28 (12.3) | 57 (25) |
| F | 20 (8.8) | 11 (4.8) | 31 (13.6) | |
| 36-59 | M | 5 (2.3) | 8 (3.5) | 15 (5.8) |
| F | 4 (1.7) | 5 (2.2) | 9 (3.9) | |
| Total N (%) | 116 (50.9) | 112 (49.1) | 228 (100) | |
Table 1: Socio‑demographic status of study participants.
M : Male F : Female n = N = Number
M : Male F : Female n = N = Number
Magnitude of Salmonella sp
The number of stools loosed per day by the 228 diarrheal children ranged from 1 to 10 stools per 24 hours. Analysis of the clinical data revealed that 183 children (80.3%) loosed 1 to 5 stools per day and only 45 children (19.7%) loosed more than 5 stools per day ie an average of 3 stools per day. These stools were fluid (65.8%), mucous (17.5%) and bloody (16.7%).
The number of stools loosed per day by the 228 diarrheal children ranged from 1 to 10 stools per 24 hours. Analysis of the clinical data revealed that 183 children (80.3%) loosed 1 to 5 stools per day and only 45 children (19.7%) loosed more than 5 stools per day ie an average of 3 stools per day. These stools were fluid (65.8%), mucous (17.5%) and bloody (16.7%).
Of 228 diarrheal children, Salmonella species was detected in 3.07% while among the control group, Salmonella was detected in 4.25% (OR 0.71 ; 95% CI 0.15-5.15). Our results showed that 33% of the Salmonella strains were isolated from patients under 12 months of age. Twenty-two (22%) of patients aged between 13-24 months of age had Salmonella and 46% of patients aged 25-59 months reported Salmonella (Table 2).
| Age (months) | Number of patients (%) | Total (%) | |
| Diarrheal group | Control group | ||
| 0-12 | 1 (14.3) | 2 (100) | 3 (33.3) |
| 13-24 | 2 (28.6) | 0 (0) | 2 (22.2) |
| 25-59 | 4 (57.1) | 0 (0) | 4 (44.5) |
| Total (%) | 7 (100) | 2 (100) | 9 (100) |
Table 2: Salmonella strains distribution according to age group.
Factors associated with diarrhea and Salmonella infection
Fever, vomiting, dehydration and anemia were associated at diarrhea respectively at 56%, 41%, 17% and 2%. Several symptoms like malnutrition (32/228; 18%) and cough (15%) were also reported in children with diarrhea. Our study reported that 3 (43%) Salmonella in diarrheal children were associated to malnutrition. Our results showed that most of the children were breastfed (58.3%) while 41.7% consumed the family dish. A few cases of herbal teas administered to the children with diarrhea were also reported (1.3%). About water supply sources, nearly half of the participants used well water as a source of drinking (47.8%). The "Office National de l’Eau et de l’Assainissement" (National Water Supply Society) and drilling water were accessible to 27.2% and 22.8% respectively. Mineral water (1.8%) and running water (0.4%) were also drunk by several diarrheal children.
Fever, vomiting, dehydration and anemia were associated at diarrhea respectively at 56%, 41%, 17% and 2%. Several symptoms like malnutrition (32/228; 18%) and cough (15%) were also reported in children with diarrhea. Our study reported that 3 (43%) Salmonella in diarrheal children were associated to malnutrition. Our results showed that most of the children were breastfed (58.3%) while 41.7% consumed the family dish. A few cases of herbal teas administered to the children with diarrhea were also reported (1.3%). About water supply sources, nearly half of the participants used well water as a source of drinking (47.8%). The "Office National de l’Eau et de l’Assainissement" (National Water Supply Society) and drilling water were accessible to 27.2% and 22.8% respectively. Mineral water (1.8%) and running water (0.4%) were also drunk by several diarrheal children.
Antimicrobials susceptibility testing
Nine (9) Salmonella sp. were isolated belonging to six different serotypes. The different rates were 22.2% for each serotype S. Typhimurium, S. Poona, S. Virchow and 11.1% for each S. Duisburg, S. Hvittingfoss, S. Ouakam. Sensitivity testing showed that the Salmonella sp strains had different levels of resistance to the antibiotics tested. Strains were resistant mainly to amoxicillin-clavulanic acid (89%), amoxicillin (100%), ceftriaxone (56%) and aztreonam (44%). We also noted resistance to quinolones (22% resistant to nalidixic acid) and fluoroquinolones (11% resistant to ciprofloxacin) (Figure 2).
Nine (9) Salmonella sp. were isolated belonging to six different serotypes. The different rates were 22.2% for each serotype S. Typhimurium, S. Poona, S. Virchow and 11.1% for each S. Duisburg, S. Hvittingfoss, S. Ouakam. Sensitivity testing showed that the Salmonella sp strains had different levels of resistance to the antibiotics tested. Strains were resistant mainly to amoxicillin-clavulanic acid (89%), amoxicillin (100%), ceftriaxone (56%) and aztreonam (44%). We also noted resistance to quinolones (22% resistant to nalidixic acid) and fluoroquinolones (11% resistant to ciprofloxacin) (Figure 2).
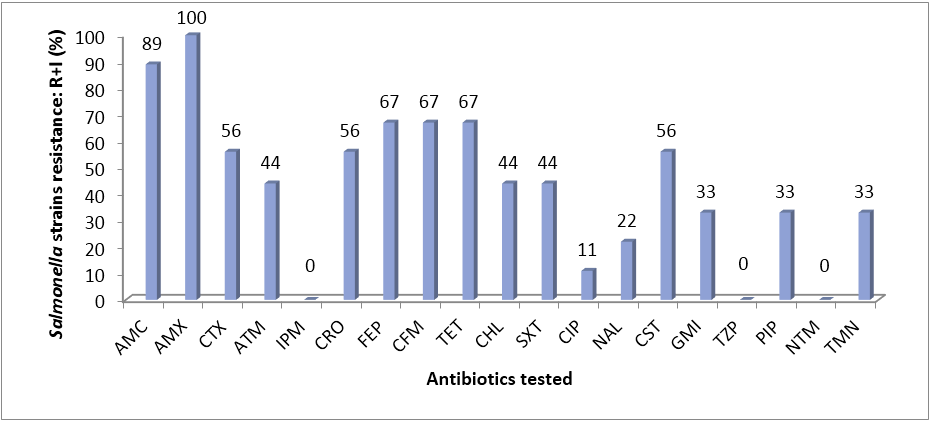
Figure 2: Resistance to individual antimicrobial among Salmonella strains.
Legend : AMC = Amoxicillin- clavulanic acid, AMX = Amoxicillin, CTX = Cefotaxime, ATM = Aztreoname, IPM = Imipenem, CRO = Ceftriaxone, FEP = Cefepime, CFM = Cefixime, TET = Tetracycline, CHL = Chloramphenicol, SXT = Trimethoprim-sulfametoxazole CIP = Ciprofloxacine, NAL = nalidixic acid, CST = Colistin sulfate, GMI = Gentamicin, TZP = Piperacillin-tazobactam, PIP = Piperacillin, NTM = Netilmicin, TMN = Tobramycin, I = Intermediate, R = Resistant.
Legend : AMC = Amoxicillin- clavulanic acid, AMX = Amoxicillin, CTX = Cefotaxime, ATM = Aztreoname, IPM = Imipenem, CRO = Ceftriaxone, FEP = Cefepime, CFM = Cefixime, TET = Tetracycline, CHL = Chloramphenicol, SXT = Trimethoprim-sulfametoxazole CIP = Ciprofloxacine, NAL = nalidixic acid, CST = Colistin sulfate, GMI = Gentamicin, TZP = Piperacillin-tazobactam, PIP = Piperacillin, NTM = Netilmicin, TMN = Tobramycin, I = Intermediate, R = Resistant.
Resistance phenotypes observed
Among the nine (9) Salmonella sp. strains, the most resistant phenotypes were Extended Spectrum β-lactamases (ESBL) phenotype (n = 4; 44.5%), Low Level Penicillinases (LLP) phenotype (n = 3; 33.3%). We also reported Low Level Penicillinases/quinolones cross-resistance phenotype (LLP/QCR) (n = 1; 11.1%) and Low Level Penicillinases/fluoroquinolones cross-resistance phenotype (LLP/FQCR) (n = 1; 11.1%) (Table 3). Multiple drug resistance (MDR) has been reported for most of the Salmonella serotypes (7/9 :78%) (Table 4).
Among the nine (9) Salmonella sp. strains, the most resistant phenotypes were Extended Spectrum β-lactamases (ESBL) phenotype (n = 4; 44.5%), Low Level Penicillinases (LLP) phenotype (n = 3; 33.3%). We also reported Low Level Penicillinases/quinolones cross-resistance phenotype (LLP/QCR) (n = 1; 11.1%) and Low Level Penicillinases/fluoroquinolones cross-resistance phenotype (LLP/FQCR) (n = 1; 11.1%) (Table 3). Multiple drug resistance (MDR) has been reported for most of the Salmonella serotypes (7/9 :78%) (Table 4).
| Antibiotic resistance phenotypes | Resistance (I+R) N (%) | ||||
| Age groups (years) | Total N (%) | ||||
| [1-2] | [2-3] | [3-4] | [4-5] | ||
| LLP | 1 (33.3) | 1 (33.3) | 1 (33.4) | 0 (0) | 3 (100) |
| ESBL | 2 (50) | 1 (25) | 1 (25) | 0 (0) | 4 (100) |
| QCR + LLP | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) |
| FQCR + LLP | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
Table 3: Distribution by age group of antibiotic resistance phenotypes of salmonella sp.
Legend: LLP = Low Level Penicillinases, ESBL = Extended Spectrum β-lactamases, QCR = Quinolones Cross-Resistance phenotype, FQCR = Fluoroquinolones Cross-Ressistance phenotype, N = number
Legend: LLP = Low Level Penicillinases, ESBL = Extended Spectrum β-lactamases, QCR = Quinolones Cross-Resistance phenotype, FQCR = Fluoroquinolones Cross-Ressistance phenotype, N = number
| Salmonella code | Serovar (Formula) | Phenotype of antibiotic resistance(a) | MDR Salmonella(b) |
| 084B | Duisburg (4, 12:d:e,n,z15) | AMC, AMX, CTX, PIP, ATM, CRO, FEP, CFM, TET, SXT, CST, GMI, TMN, ESBL | Yes |
| 057B | Poona (13, 22:z:1,6) | AMC, AMX, FEP, TET, CHL, CST, GMI, LLP | Yes |
| 066B | Typhimurium (4,5, 12:i:1,2) | AMC, AMX, TET, SXT, CIP, NAL, GMI, LLP, FQCR | Yes |
| 068B | Typhimurium (4,5, 12:i:1,2) | AMX, CTX, ATM, CRO, FEP, CFM, SXT, CST, ESBL | Yes |
| 078B | Ouakam (9, 46:z29:-) | AMC, AMX, TET, CST, TMN, LLP | Yes |
| 063G | Hvittingfoss (16:b:e,n,x) | AMC, AMX, CTX, ATM, CRO, FEP, PIP, CFM, CHL, ESBL | No |
| 087G | Poona (13, 22:z:1,6) | AMC, AMX, CTX, ATM, PIP, CRO, FEP, CFM, TET, CHL, CST, TMN, ESBL | Yes |
| 112G1 | Virchow (6, 7:r:1,2) | AMC, AMX, CTX, CRO, FEP, CFM, TET, SXT, NAL, CHL, LLP, QCR | Yes |
| 112G2 | Virchow (6, 7:r:1,2) | AMC, AMX, CFM, LLP | No |
Table 4: Phenotypic antibiotic resistance patterns of Salmonella serovars isolated.
(a)Antibiotic resistance patterns of Salmonella. AMC, Amoxicillin- clavulanic acid ; AMX, Amoxicillin ; CTX, Cefotaxime ; ATM, Aztreoname ; CRO, Ceftriaxone ; FEP, Cefepime ; CFM, Cefixime ; TET, Tetracycline ; CHL, Chloramphenicol ; SXT, Trimethoprim-sulfametoxazole ; CIP, Ciprofloxacine ; NAL, nalidixic acid ; CST, Colistin sulfate ; GMI, Gentamicin ; PIP, Piperacillin ; TMN, Tobramycin ; LLP, Low Level Penicillinases; ESBL, Extended Spectrum β-lactamases; QCR, Quinolones Cross-Resistance phenotype; FQCR, Fluoroquinolones Cross-Resistance phenotype.
(b)MDR : Multidrug resistance
(a)Antibiotic resistance patterns of Salmonella. AMC, Amoxicillin- clavulanic acid ; AMX, Amoxicillin ; CTX, Cefotaxime ; ATM, Aztreoname ; CRO, Ceftriaxone ; FEP, Cefepime ; CFM, Cefixime ; TET, Tetracycline ; CHL, Chloramphenicol ; SXT, Trimethoprim-sulfametoxazole ; CIP, Ciprofloxacine ; NAL, nalidixic acid ; CST, Colistin sulfate ; GMI, Gentamicin ; PIP, Piperacillin ; TMN, Tobramycin ; LLP, Low Level Penicillinases; ESBL, Extended Spectrum β-lactamases; QCR, Quinolones Cross-Resistance phenotype; FQCR, Fluoroquinolones Cross-Resistance phenotype.
(b)MDR : Multidrug resistance
Discussion
In this study, the proportion of detected Salmonella was similar to the finding from others previous worldwide studies : 4% [20], Ethiopia (Addis Ababa), 3.95% [21], Ethiopia (Butajira), 4.5% [22]. In addition, our result was in line with the study conducted in Nepal [23], Turkey [24] where the detection of Salmonella were 4.6 and 3 respectively.
The isolation rate of Salmonella from this study was found to be lower than 9%, previously reported in Ouagadougou [25], 6.2% reported in Jimma [26], 12.6% in South Ethiopia [27]. This was unexpected since hygiene and sanitation are better in urban than rural areas and thus may reduce Salmonella incidence. However, some practices like those that street food system that is more developed in Ouagadougou may justify the high prevalence in urban area. This is consistent with Ameya., et al. [27] who found that urban dwellers were more infected with the enteric pathogen than rural residents.
On the other hand, the prevalence of the Salmonella found from study was higher than those obtained in Ethiopia : 1% from Abebe., et al. [28] and 1.5% from Getamesay., et al. [29]. This could probably be explained by different factors known to influence the bacteria prevalence such as : the study population, study period, geographical and seasonal variation but also the increased of the community awareness about personal and environmental hygiene. Indeed, a better awareness of the community especially mothers directly influence the prevalence of Salmonella among the children [28]. The variation may also be due to the socio-economic status, source of drinking water supply, sanitation and hygiene practices of the people in the cites [27].
In fact, our results showed that most of the children who participated in this study were breastfed. In general, the most common mode of feeding in rural areas is breastfeeding until the weaning period is reached. The breastfeeding process could be the cause of some contamination by enteropathogens, as the breasts are usually not cleaned before being given to the child. In addition, from 4 to 5 months of age, breast milk alone becomes insufficient to adequately ensure the growth of the child. Then additional nutriments to the milk are required. Unfortunately, this nutritional supplement, if not well treated, may expose the child to certain enteropathogens and therefore to gastroenteritis. In most of the cases, because of the low income of rural populations, the family dish has a low nutritional value. This could then expose the children susceptibility to diarrheal diseases because children living in poor areas with poor hygiene and sanitation conditions and children with poor nutritional status are most at risk of developing persistent diarrhoea [30, 20]. As poor nutrition is both a risk factor and a consequence of persistent diarrhoea, both are very commonly associated [20]. Otherwise, in rural settings, mothers sometimes refer to herbal teas and other decoctions when the children were diarrhea affected. Indeed previous study showed that 14% of diarrheal children used traditional medicine (herbal tea or various natural powders mixed with water) as heath care, which can even be contaminated with pathogens causing Gastrointestinal Infections (GI), thus being harmful them [31].
The present study also revealed that nearly half of the participants used well water as a source of drinking. In most cases, water supply sources could be reservoirs and then be vectors of transmissions of enteric pathogens, water being a risk factor for infectious diseases [32].
Among the nine Salmonella isolates, the overall rate of resistance was high for aminopenicillins [amoxicillin-clavulanic acid (89%), amoxicillin (100%)] and monobactam [aztreonam (44%)] which is comparable with results of a study, where amoxicillin (100%) were reported in Ethiopia [33,34]. Elevated resistances were noted to 3rd generation cephalosporins (ceftriaxone) which is in agreement with the study of Mulatu., et al. [34] where resistance to ceftriaxone (75%) was reported. Resistances to quinolones (22% to nalidixic acid) and fluoroquinolones (11% to ciprofloxacin) were also shown. Our findings are similar a study where 25% of Salmonella isolates were resistance to nalidixic acid [34]. Contrary to our findings, a high level of antibiotic susceptibility of Salmonella to chloramphenicol, ciprofloxacin and norfloxacin was shown in Ethiopia [26,28]. The rise in resistance might be due to the selective pressure created by the use of antimicrobials in food processing animals and irrational use of antibiotics [35].
As for most of the isolated serotypes, S. Typhimurium was found to be resistant to eight antibiotics (AMC, AMX, TET, SXT, CIP, NAL, GMI, LLP and AMX, CTX, ATM, CRO, FEP, CFM, SXT, CST), results similar to MDR S. Typhimurium isolated from human, chicken and cattle from Malaysia and Colombia [36,37]. MDR phenotype of S. Duisburg, S. Poona, S. Virchow, S. Typhimurium and S. Hvittingfoss isolated in this study seems similar to thesse reported from chicken carcasses [37] indicating an increased risk and concern in the possible contamination of humans through animals.
Antibiotic treatment of salmonellosis is complicated because microorganism under antibiotic pressure may select for virulence within the host [38], acquires tolerance and multiple drug resistance (MDR) phenotypes (fast-, moderate- and low-growing subsets) within host tissues [39] and frequently incorporate new genetic material to resist to the antibiotic selective pressure [40].
Conclusion
This study found that about 80% of Salmonella serovars isolated from children with and without diarrhea in rural settings of Burkina Faso were multidrug-resistant. Data are consistent with the global health concern imposed by antibiotic resistant strains that may limit the choice of treatment of human infections. Our study provided valuable information on risk factors associated with children diarrhea in Burkina Faso. Results also suggest various needs that include cooperation between the sector of stock farming, governmental and academic institutions to improve surveillance of both Salmonella and its antibiotic resistance patterns in broiler farms, poultry products and humans. To reach this aim, it would be suitable to establish appropriate regulations and funding for antibiotic research, and to promote education and prudent use of antibiotics as well as by clinicians like farmers.
Acknowledgements
The authors gratefully thank the "Laboratoire de Biologie Moléculaire, d’Épidémiologie et de Surveillance des Bactéries et Virus Transmissibles par les Aliments (LaBESTA)", the "Centre National de Recherche et de Formation sur le Paludisme (CNRFP)" for their technical support. We also thank the parents and guardians of children as well as the authorities of the "CMA de Boromo" and "CMA de Gourcy" for their kind cooperation.
The authors gratefully thank the "Laboratoire de Biologie Moléculaire, d’Épidémiologie et de Surveillance des Bactéries et Virus Transmissibles par les Aliments (LaBESTA)", the "Centre National de Recherche et de Formation sur le Paludisme (CNRFP)" for their technical support. We also thank the parents and guardians of children as well as the authorities of the "CMA de Boromo" and "CMA de Gourcy" for their kind cooperation.
Conflict of interest
The authors declare that they have no conflict interests.
The authors declare that they have no conflict interests.
References
- Kosek M., et al. "The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000". Bulletin of the World Health Organization 81.3 (2003): 197-204.
- World Health Organization (WHO). Diarrheal disease. 2015.
- World Health Organization (WHO). Diarrhea ; Why children are still dying and what can be done. 2009.
- Nguyen Vu T., et al. "Etiology and epidemiology of diarrhea in children in Hanoi, Vietnam". International Journal of Infection 10.4 (2006): 298–308.
- Fashae K., et al. "Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria". The Journal for Infection in Developing Countries 4.8 (2010): 484-494.
- Shi Q., et al. "Situation of Salmonella contamination in food in Hebei, Province of China in 2009 -2010". African Journal of Microbiology Research 6.2 (2012): 365-370.
- Kirk MD., et al. "World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010 : A data synthesis". PLoS Med 12 (2015): e1001921.
- Imbert P. "Prise en charge des diarrhées aiguës de l’enfant en milieu tropical". Médecine Tropicale 61 (2001) : 226–230.
- World Health Organization (WHO). "Antimicrobial Resistance: Global Report on Surveillance" (2014).
- Gordon MA., et al. "Invasive non-typhoid salmonellae establish systemic intracellular infection in HIV-infected adults: an emerging disease pathogenesis". Clinical Infectious Diseases 50 (2010): 953-962.
- Sire JM., et al. "Low-level resistance to ciprofloxacin in non-Typhi Salmonella enterica isolated from human gastroenteritis in Dakar, Senegal (2004-2006)". International Journal of Antimicrobial Agents 31.6 (2008) : 581-582.
- Hendriksen RS., et al. "Emergence of Multidrug-Resistant Salmonella Concord Infections in Europe and the United States in Children Adopted From Ethiopia, 2003-2007". The Pediatric Infectious Disease Journal 28 (2009): 814-818.
- Urio EM., et al. "Shigella and Salmonella strains isolated from children under 5 years in Gaborone, Botswana, and their antibiotic susceptibility patterns". Tropical Medicine and International Health 6.1 (2001): 55-59.
- Moyo SJ., et al. "Age specific aetiological agents of diarrhoea in hospitalized children aged less than five years in Dar es Salaam, Tanzania". BMC Pediatrics 11 (2011): 19.
- Popoff MY., et al. "Supplement 2002 (no. 46) to the Kauffmann-White scheme". Research in Microbiology 155.7 (2004): 568-570.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). "Recommandation 2017". 1.0 (2017): 1-127.
- Guessennd N., et al. "Prévalence et profil de résistance des entérobactéries productrices de β-lactamases à spectre élargi (BLSE) à Abidjan côte d’ivoire de 2005 à 2006". Journal of Science and Pharmeucical Biology 9 (2008): 63-70.
- Philippon A., et al. "Entérobactéries et β-lactamines: phénotypes de résistance naturelle". Pathologie Biologie 60.2 (2012): 112-26.
- Kamga HG., et al. "Phénotypes de résistance des souches d’Escherichia coli responsables des infections urinaires communautaires dans la ville de Yaoundé (Cameroun)". African Journal Pathology and Microbiology 3 (2014): 1-4.
- Abba K., et al. "Pathogens associated with persistent diarrhea in children in low and middle income countries: systematic review". The Journal Infectious Diseases 9 (2009): 88.
- Mamuye Y., et al. "Isolation and antibiotic susceptibility patterns of Shigella and Salmonella among under 5 children with acute Diarrhoea: a cross-sectional study at selected public health facilities in Addis Ababa, Ethiopia". Clinical Microbiology: Open Access 4 (2015): 186.
- Mengstu G., et al. "Prevalence and antimicrobial susceptibility patterns of Salmonella serovars and Shigella species". Journal of Microbial and Biochemical Technology 2 (2014): 1–7.
- Ansari S., et al. "Bacterial etiology of acute diarrhea in children under five years of age". Journal of Nepal Health Research Council 10.22 (2012) :218–223.
- Kara TT.,et al. "Prevalence of Salmonella and Shigella spp. and antibiotic resistance status in acute childhood gastroenteritis". Journal of Pediatric Infections 9 (2015): 102–107.
- Bonkoungou IJO., et al. "Bacterial and viral etiology of childhood diarrhea in Ouagadougou, Burkina Faso". BMC Pediatrics 13 (2013): 36 doi: 10.1186/1471-2431-13-36.
- Beyene G and Tasew H. "Prevalence of intestinal parasite, Shigella and Salmonella species among diarrheal children in Jimma health center, Jimma southwest Ethiopia: a cross sectional study". Annal of Clinical Microbiology and Antimicrobials13 (2014) :10.
- Ameya G., et al. "Antimicrobial susceptibility pattern, and associated factors of Salmonella and Shigella infections among under five children in Arba Minch, South Ethiopia". Annal of Clinical Microbiology and Antimicrobials 17.1 (2018): 1.
- Abebe W., et al. "Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with Diarrhoea attending Nigist Eleni Mohammed memorial hospital, South Ethiopia". BMC Pediatrics 18 (2018) :241.
- Getamesay M., et al. "Prevalence of Shigella, Salmonella and Campylobacter species and their susceptibility pattern among under five children with diarrhea in Hawassa town, South Ethiopia". Ethiopian Journal of Health Science 24.2 (2014) :101–108.
- Lima AAM and Guerrant RL. "Persistent diarrhea in children : epidemiology, risk factors, pathophysiology, nutritional impact, and management". Epidemiologic Reviews 14 (1992): 222-242.
- Dembélé R., et al. "Burden of acute gastrointestinal infections in Ouagadougou, Burkina Faso". Journal of Microbiology and Infectious Diseases 6.2 (2016): 45-52.
- Morpeth SC., et al. "Invasive Non-Typhi Salmonella Disease in Africa". Clinical Infectious Diseases49 (2009): 607-611.
- Reda A., et al. "Antibiotic susceptibility patterns of Salmonella and Shigella isolates in Harar, eastern Ethiopia". Journal of Infectious Diseasees and Immunology 3.8 (2011): 134–139.
- Mulatu G., et al. "Prevalence of Shigella, Salmonella and Campylobacter species and their susceptibility patters among under five children with diarrhea in Hawassa town, South Ethiopia". Ethiopian Journal of Health Science 24.2 (2014): 101–108.
- Addisalem HB., et al. "Tetracycline residue levels in slaughtered beef cattle from three slaughterhouses in Central Ethiopia". Journal of Global Veterinary 8.6 (2012): 546–554.
- Benacer DTKL., et al. "Characterization of drug resistant Salmonella enterica serotype Typhimurium by antibiograms, plasmids, integrons, resistance genes and PFGE". Journal of Microbiology and Biotechnology 20.6 (2010): 1042–1052.
- Vélez CD., et al. "Phenotypic and Genotypic Antibiotic Resistance of Salmonella from Chicken Carcasses Marketed at Ibague, Colombia". Brazilian Journal of Poultry Science 19.2 (2017): 347-354.
- Diard M., et al. "Antibiotic treatment selects for cooperative virulence of Salmonella typhimurium". Current Biology 24.17 (2014): 2000–2005.
- Claudi B., et al. "Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy". Cell 158.4 (2014): 722–733.
- Brown-Jaque M., et al. "Transfer of antibioticresistance genes via phage-related mobile elements". Plasmid 79 (2015): 1–7.
Citation:
René Dembélé., et al. “Prevalence of Multidrug-resistant Salmonella enterica and associated factors among under five children
with diarrhea in rural Burkina Faso.” Clinical Biotechnology and Microbiology 3.1 (2018): 566-576.
Copyright: © 2018 René Dembélé., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.






























 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.