Research Article
Volume 2 Issue 1 - 2018
Inhibition of Insulin Receptor Tyrosine Kinase by the Firmicutes Metabolome during Insulin Resistance
1Centre for Biocomputing and Drug Development, Adekunle Ajasin University, Nigeria
2National Biotechnology Development Agency, Abuja, Nigeria
3Department of Biochemistry, Adekunle Ajasin University, Nigeria
2National Biotechnology Development Agency, Abuja, Nigeria
3Department of Biochemistry, Adekunle Ajasin University, Nigeria
*Corresponding Author: Adewale J Ogunleye, Centre for Biocomputing and Drug Development, Adekunle Ajasin University, Nigeria.
Received: August 17, 2018; Published: August 24, 2018
Abstract
A putative mechanism for the disruption of insulin signalling via inhibition of the intrinsic phosphorylation feature of insulin receptor tyrosine kinase (IRTK) by primary and secondary metabolites of firmicutes was described in this paper. An in-silico pipeline was employed for the first time, to screen for the upregulated metabolic signatures and pathways of firmicutes in the gut, serum and plasma of type 2 diabetes mellitus (T2DM) patients. It was realized that (i) metabolic exudates of the biosynthesis of secondary bile acids (SBAs) by microbes and not humans had a high binding affinity towards IRTK and (ii) secondary metabolites produced by microbes during the production of branched chain amino acids (BCAA) have a high binding affinity for IRTK, and not BCAAs themselves. These results, although putative, provide a clear understanding of the overall contribution of firmicutes to insulin resistance, an important hallmark of T2DM.
Keywords: Human gut micro biome; Metabolomics; Insulin receptor tyrosine kinase (IRTK); Type 2 diabetes mellitus (T2DM); Secondary Bile Acids (SBA); Branched Chain Amino Acids (BCAA)
Introduction
Type II Diabetes mellitus (T2DM) is a chronic metabolic disorder with one of the greatest disease burdens recorded globally [1]. As at 2014, the WHO reports that 422million adults (8.5% of adult population) suffer from diabetes globally. This is as nearly double its prevalence in 1980 (4.7% of adult population). Just around late 2017, an estimate reports that over 1 in 11 adults suffer from diabetes, of which 90% of them suffer from T2DM [2]. This confirms that diabetes (especially T2DM) is on the rise, and at the same time, a reflection on the insufficiency of current anti-diabetic efforts.
In addition to dysfunctional displays by the pancreatic islets, insulin resistance (INSR) is another established hallmark of T2DM that best predicts and sustains chronic hyperglycaemia [3-7]. With the sole aim of mopping up excess glucose, insulin initiates its downstream signalling effects by binding to the extracellular domain of its receptor [7-9]. Insulin binding triggers the phosphorylation of insulin receptor (IR) on the tyrosine core of its intracellular kinase domain. This subsequently leads to the recruitment and phosphorylation of special adaptor proteins known as insulin receptor substrates 1/2 (IRS-1/2). IRS-1/2 are important signalling intermediates that are; (I) directly phosphorylated by the tyrosine kinase subunit of insulin Receptor (II) involved in the activation of phospotidylinositol-3-kinase (PI3K) and Protein Kinase B (PKB/AKT) pathways. These steps are crucial in potentiating the vesicular release of glucose transporter 4 (GLUT4) which subsequently internalizes extracellular glucose [7-10]. Throughout the whole insulin signalling pathway, an abnormal functioning IRTK (Figure 1) has been identified to play a central role in insulin resistance [11-12]. Simply put, the blockade of this enzyme means that no insulin signals are transduced, even if insulin is produced in excess.
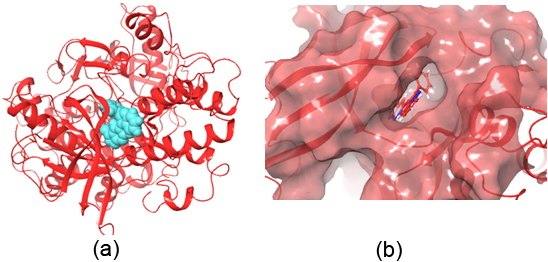
Figure 1: Crystal structure of the Insulin Receptor) bound to Irfin-1 (turquoise, PDB-1D: 4IBM; ΔG = -7.756 kcal/mol). (b) Surface representation of Irfin-1 within the insulin receptor’s tyrosine kinase site.
Recently, scientific evidences have identified Firmicutes as a distinct microbiome signatures that clearly defines T2-diabetic individuals from persons with normal glucose tolerance (NGT) [13-16]. Although the direct effect of these microbes is unknown, so many propositions have been made. For example, microbes are linked to increase in Glucagon-like peptides (GLP-1/2) secretion; a step necessary for the release of insulin, stimulation of postprandial release of peptides associated with pancreatic functions etc. [13,16,17]. From all these studies, it is clear that T2DM-specific microbes interact with the gastrointestinal lumen of diabetics via their metabolome therefore leading to insulin resistance [3,13,17]. However, the direct proteo-metabolome interaction which makes firmicutes contribute to insulin resistance is unknown. The present study seeks to answer this question, and in a way, proffer probable insights into how metabolic exudates from firmicutes in T2DM patients suppresses IRTK; a critical node downstream insulin signalling.
Results and Discussion
In recent times, the contribution of the microbiome and their metabolites to the pathogenesis of non-communicable diseases is becoming an area of active research. Previous studies have shown that a constitutional shift must occur in the tissue microbiome before and during disease development [3,5]. In the case of diabetes, studies have shown that firmicutes (as an Operational Taxonomic Unit; OTU) are prominent during chronic T2DM and not in normoglycemic persons [13,17]. As suggested by Gonzalez-Fransquesa [3], we utilized an in-silico approach (molecular docking) to provide a feasible, yet simple understanding of insulin intolerance.
Molecular Docking
Molecular docking was performed to predict the binding affinity of these metabolites relative to irfin-1 (ΔG = -7.756 kcal/mol), a known inhibitor of the kinase domain of insulin receptor. It is known that IRTK contains a tyrosine rich sub site called phospho-tyrosine (pTyr). The inhibition of this site abolishes downstream insulin signalling, and it has been a proven strategy for inhibiting IRTK [18]. This site is also responsible for the intrinsic phosphorylation feature of the insulin receptor [8, 19-21]. Hence, all docking calculations and grid definitions were centred on the pTyr sub site. Figure 2 is a heatmap representing the docking score of all the metabolites that were screened. From the docking scores, the metabolite can be compared with irfin-1 on the basis of inhibition. In fact, the higher docking results suggests that in the presence of metabolically active firmicutes, the metabolites have a higher potential to inhibit phosphorylation of IRTK.
Molecular docking was performed to predict the binding affinity of these metabolites relative to irfin-1 (ΔG = -7.756 kcal/mol), a known inhibitor of the kinase domain of insulin receptor. It is known that IRTK contains a tyrosine rich sub site called phospho-tyrosine (pTyr). The inhibition of this site abolishes downstream insulin signalling, and it has been a proven strategy for inhibiting IRTK [18]. This site is also responsible for the intrinsic phosphorylation feature of the insulin receptor [8, 19-21]. Hence, all docking calculations and grid definitions were centred on the pTyr sub site. Figure 2 is a heatmap representing the docking score of all the metabolites that were screened. From the docking scores, the metabolite can be compared with irfin-1 on the basis of inhibition. In fact, the higher docking results suggests that in the presence of metabolically active firmicutes, the metabolites have a higher potential to inhibit phosphorylation of IRTK.

Figure 2: Heatmap representation of the docking score of each metabolite. Each of the metabolites were grouped into cholate, methylamine, pyruvate and uracil scaffolds.
Secondary bile acids (SBA) have a high inhibitory affinity for IRTK
By way of molecular docking, it was observed that compounds with the cholate scaffold had a high binding affinity towards IRTK. Upon KEGG pathway enrichment, it was verified that these compounds were biosynthetic products of secondary bile acid (SBA) pathway. Also the results shown in figure 4 reveals that microbe-derived SBAs and not human-derived SBAs specifically inhibit IRTK. Such microbe-derived SBA metabolites include sodium cholic acid (-8.949 kcal/mol), alpha-phocaecholic (-8.289 kcal/mol) beta-phocaecholic acid (-8.173 kcal/mol), Chenodeoxycholic acid, (-8.118 kcal/mol), Allochenodeoxycholic acid (-8.042 kcal/mol) and Haemulcholic acid (-8.036 kcal/mol). (See full docking result on table 1). Upon ligand interaction analysis, (shown in figure 3) hydrogen bonding between the metabolites and one or more of Met1079, Arg1000, Ala1080 and Tyr1011 were observed. With the aid of these hydrogen contacts, the hydrophobic ends of SBAs were completely buried within the pocket while their hydrophilic ends experience solvent exposure.
By way of molecular docking, it was observed that compounds with the cholate scaffold had a high binding affinity towards IRTK. Upon KEGG pathway enrichment, it was verified that these compounds were biosynthetic products of secondary bile acid (SBA) pathway. Also the results shown in figure 4 reveals that microbe-derived SBAs and not human-derived SBAs specifically inhibit IRTK. Such microbe-derived SBA metabolites include sodium cholic acid (-8.949 kcal/mol), alpha-phocaecholic (-8.289 kcal/mol) beta-phocaecholic acid (-8.173 kcal/mol), Chenodeoxycholic acid, (-8.118 kcal/mol), Allochenodeoxycholic acid (-8.042 kcal/mol) and Haemulcholic acid (-8.036 kcal/mol). (See full docking result on table 1). Upon ligand interaction analysis, (shown in figure 3) hydrogen bonding between the metabolites and one or more of Met1079, Arg1000, Ala1080 and Tyr1011 were observed. With the aid of these hydrogen contacts, the hydrophobic ends of SBAs were completely buried within the pocket while their hydrophilic ends experience solvent exposure.
| Compound | Docking Score |
| Cholic acid | -8.953 |
| Alpha-phocaecholic acid | -8.289 |
| Phocaecholic acid | -8.730 |
| Chenodeoxycholic acid | -8.800 |
| Allochenodeoxycholic acid | -8.042 |
| Allodeoxycholate | -5.361 |
| Avicholic acid | -5.289 |
| Haemulcholic acid | -8.036 |
| Taurodeoxycholate | -7.635 |
| Deoxycholic acid | -7.628 |
| Bitocholic acid | -7.495 |
| Alloavicholic acid | -7.453 |
| Lithocholic acid | -7.413 |
| Glycochenodeoxycholic acid | -7.364 |
| Hyocholic acid | -7.301 |
| Gamma-muricholate | -7.301 |
| Ursocholic acid | -7.294 |
| Cygnocholic acid | -7.259 |
| Avideoxycholic acid | -7.057 |
| 12-epideoxycholate | -3.894 |
| Alpha-muricholate | -3.704 |
| Avicholate | -6.708 |
| Avideoxycholate | -6.981 |
| Beta-muricholate | -3.496 |
| Chenodeoxycholoyl-CoA | -4.361 |
| Cholate | -8.949 |
| Glycochenodeoxycholate | -7.364 |
| Glycocholate | -7.110 |
| Hyocholate | -7.263 |
| Hyodeoxycholate | -4.263 |
| Isochenodeoxycholate | -4.723 |
| Isolithocholate | -3.322 |
| Murideoxycholate | -4.325 |
| Omega-muricholate | -5.325 |
| Taurochenodeoxycholate | -6.235 |
| Taurocholate | -6.435 |
| Ursocholate | -4.926 |
| Vulpecholate | -3.244 |
| M-cresol | -7.486 |
| Cysteine | -7.487 |
| Phenylalanine | -6.605 |
| 2-hydroxyglutaric acid | -6.910 |
| Tyramine | -6.815 |
| Valine | -6.751 |
| Hydroxyparaconate | -7.955 |
| L-pipecolic acid | -7.567 |
| L-proline | -7.387 |
| Hypoxanthine | -7.484 |
| L-glutamic acid | -4.850 |
| 1-methyluric acid | -7.622 |
| 1-methyluric acid | -7.522 |
| Leucine | -6.669 |
| Cys-gly | -6.968 |
| Kynurenine | -6.666 |
| N,n-dimethylglycine | -6.543 |
| Dimethylamine | -6.348 |
| Phenylacetylglycine | -6.880 |
| Glycine | -6.510 |
| Isoleucine | -6.480 |
| Methylamine | -6.390 |
| L-tyrosine | -5.900 |
| L-homoserine | -5.817 |
| Hydroxyparaconate | -7.955 |
| Cis-aconitic acid | -7.937 |
| Citraconic acid | -7.714 |
| Acetoin | -6.921 |
| (2r)-2-hydroxypentanedioic acid | -6.910 |
| Pyruvate | -6.806 |
| Itaconic acid | -4.795 |
| Malic acid | -7.024 |
| Isocitric acid | -7.010 |
| Salicyluric acid | -6.397 |
| Malonic acid | -6.526 |
| Taurine | -6.392 |
| 1-methyluric acid | -8.643 |
| Hippuric acid | -7.313 |
| 3-indoleacetonitrile | -7.289 |
| 2935-35-5 | -7.272 |
| 1-methyluric acid | -7.622 |
| Hypoxanthine | -7.484 |
| N-methylnicotinate | -6.991 |
| Methyl nicotinate | -6.730 |
| Kynurenine | -6.666 |
| Trimethylamine n-oxide_2 | -6.616 |
| Nicotinuric acid | -5.733 |
| Salicyluric acid | -6.749 |
| Uracil | -8.269 |
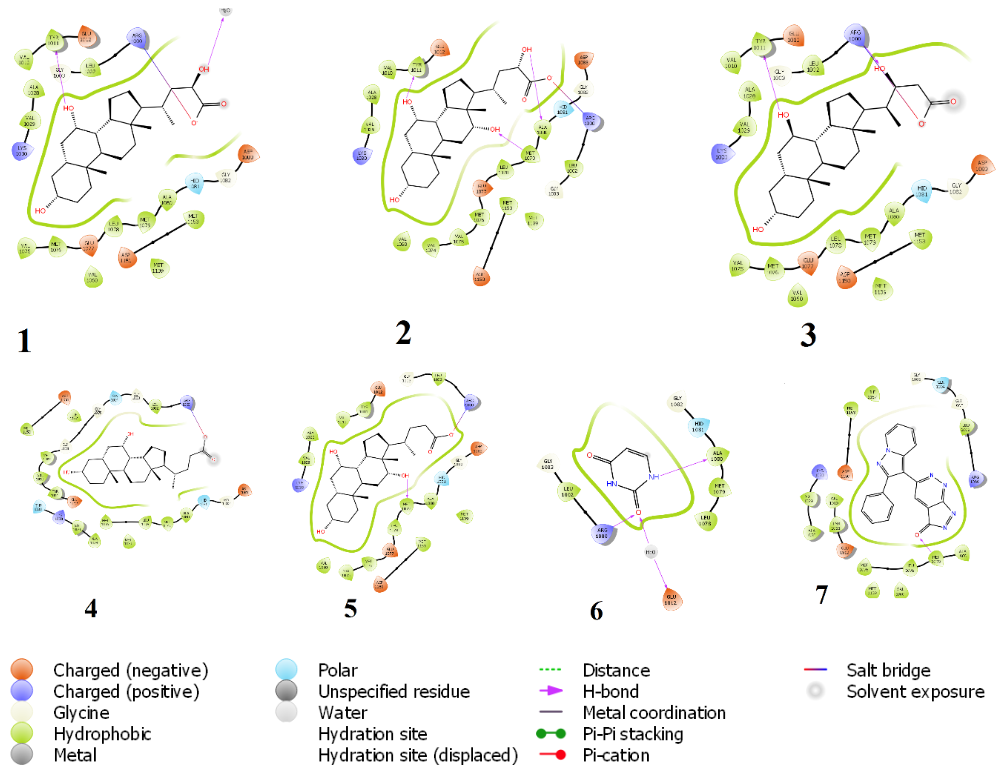
Figure 3: Chemical interaction diagram between IRTK and (1) Beta-phocaecholic acid (2) alpha phocaecholic acid (3) Haemulcholic acid (4) chenodeoxycholic acid, (5) cholic acid (6) uracil (7) irfin-1. Hydrogen bonding with either Met1079 or Arg1000 were prominent interactions for inhibition in all.
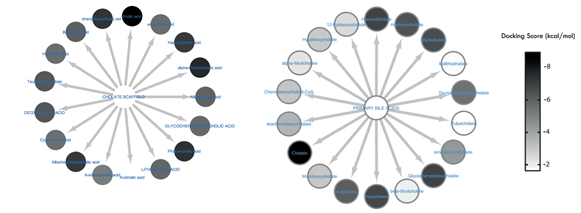
Figure 4: Comparative visualization of Secondary bile acids biosynthesized by (a) microbes and (b) humans. The docking scores were colour-mapped onto each metabolite. It is obvious that apart from avicholate, all other SBA metabolites produced by microbes and not humans had a high binding affinity for IRTK.
In two independent studies by Suhre., et al. and Zhao., et al. [22-23] , it was observed that there was a transition in the circulating pool of bile acids from primary to secondary within the serum of T2D patients. This biotransformative process was linked to four major microbial-phyla which includes; Firmicutes, Actinobacteria, Bacteroidetes and Proteobacteria. To the best of knowledge, they are the only non-vertebrates that possess the required enzymes for bio-transforming PBAs to SBAs [17, 22-23 ]. These observation and as shown by the results, the continued synthesis of microbial-SBAs can directly inhibit IRTK and could be one of the reasons for microbiome-induced insulin resistance. Figure 5 shows the lipophilic efficiency of these metabolites (as calculated by glide). Coupled with their amphipathic nature, SBAs have the ability to traverse cell membrane, thereby having access to IRTK intracellular domain.
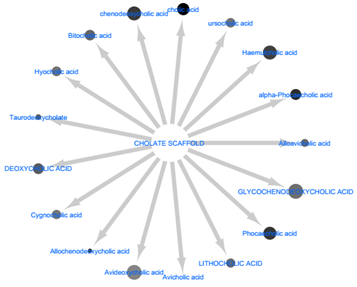
Figure 5: The relative lipophilicity of the secondary bile acids was size-mapped onto node size of each metabolite. The lipophilic efficiency is a measure of how well they can permeate phospholipid bilayer membranes. Glycohenodeoxycholc acid had the highest rate of permeability while avicholic acid had the lowest.
Firmicutes utilize multiple pathways to drive the production of branched chain amino acids (BCAA)
Metabolites from firmicutes such as uracil (-8.269 kcal/mol), citraconate (-7.714), cis-aconitate (-7.937), lactate (-7.701 kcal/mol), Phenethylamine (-7624 kcal/mol), nicotinurate (-6.991 kcal/mol), pipecolate (-7.567), 5-oxoproline (-7.620 kcal/mol) Hydroxyparaconate (-7.955 kcal/mol) and praline (-7.387 kcal/mol) points to a pathway crosslink between amino acid degradation, nucleotide sugar synthesis and niacin metabolism which is seen in the C5-branched dibasic acids metabolism pathway24. Based on the docking scores (see table 1), most of the metabolites here had a strong inhibitory potential towards IRTK. The end-product of these pathways are the BCAAs (valine, leucine and isoleucine) which have been frequently implicated as upregulated metabolite in T2D patients. The pathway (Figure 6) was obtained from KEGG.
Metabolites from firmicutes such as uracil (-8.269 kcal/mol), citraconate (-7.714), cis-aconitate (-7.937), lactate (-7.701 kcal/mol), Phenethylamine (-7624 kcal/mol), nicotinurate (-6.991 kcal/mol), pipecolate (-7.567), 5-oxoproline (-7.620 kcal/mol) Hydroxyparaconate (-7.955 kcal/mol) and praline (-7.387 kcal/mol) points to a pathway crosslink between amino acid degradation, nucleotide sugar synthesis and niacin metabolism which is seen in the C5-branched dibasic acids metabolism pathway24. Based on the docking scores (see table 1), most of the metabolites here had a strong inhibitory potential towards IRTK. The end-product of these pathways are the BCAAs (valine, leucine and isoleucine) which have been frequently implicated as upregulated metabolite in T2D patients. The pathway (Figure 6) was obtained from KEGG.
It shows that pyruvate was sourced from glycolysis, citric acid cycle, Niacin metabolism, nucleoside synthesis and amino acid degradation pathways. These pathways provide the pyruvate needed by the C5-branched dibasic acids metabolism pathway [24]. Then, pyruvate is converted to acetolactate, an essential raw material for the production of BCAAs [24]. The non-essentiality of BCAAs to firmicutes in the presence of raw materials ensures continuous production of BCAAs, while its essentiality to humans keep them circulating in the blood stream. This is an apparent reason for the over-abundance of branched chain amino acids in the serum of patients with type 2 diabetes mellitus. Given the above, it can be said in this context, that BCAAs are overexpressed biomarkers and not direct inhibitors of insulin signalling via IRTK inhibition. However, this not nullify their involvement in triggering other associated hallmarks of T2DM.
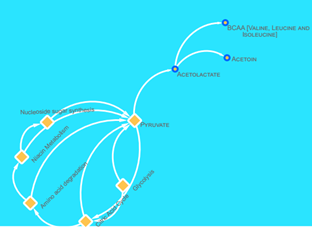
Figure 6: A brief schematic of the C5 dibasic acid biosynthesis pathway. Here, pyruvate, was either a metabolic product or substrate of nucleoside sugar synthesis, niacin metabolism, amino acid degradation, citric acid cycle and glycolysis. Pyruvate was converted to acetolactate, a raw material for the production of BCAA.
Methods
Molecular docking was performed to study the effect of metabolites generated by firmicutes on an essential insulin signalling protein, IRTK. Briefly, a manual literature search for all metabolites of firmicutes [as an Operational Taxonomic Unit (OTU)] residing in the gastrointestinal tract of type 2 diabetic patients was performed on PubMed (http://www.pubmed.ncbi.nlm.nih.gov). The major search text ‘Type 2 Diabetes AND Firmicutes AND Metabolome’. Articles with metabolomics data from 2 or more non-distinct phyla were disregarded. Using Schrodinger (Schrodinger Inc., LLC), the respective 3-D conformers of each metabolite (ligand) was generated using LigPrep. Also, inhibitor bound crystal structures of IRTK [18] (PDB: 4IBM) was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb/explore/explore.do?structureId=4IBM).
Molecular docking was performed to study the effect of metabolites generated by firmicutes on an essential insulin signalling protein, IRTK. Briefly, a manual literature search for all metabolites of firmicutes [as an Operational Taxonomic Unit (OTU)] residing in the gastrointestinal tract of type 2 diabetic patients was performed on PubMed (http://www.pubmed.ncbi.nlm.nih.gov). The major search text ‘Type 2 Diabetes AND Firmicutes AND Metabolome’. Articles with metabolomics data from 2 or more non-distinct phyla were disregarded. Using Schrodinger (Schrodinger Inc., LLC), the respective 3-D conformers of each metabolite (ligand) was generated using LigPrep. Also, inhibitor bound crystal structures of IRTK [18] (PDB: 4IBM) was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb/explore/explore.do?structureId=4IBM).
The protein preparation wizard implemented Epik (for buffering at pKa = 7.0 ± 2.0) and Prime (to fill missing loops and identify residue/atomic overlaps). The OPLS_2005 force field was used throughout to. The Glide Standard Precision (SP) scoring function was implemented for the docking of each metabolite within a pre-defined receptor grid which corresponds to the protein’s active site. The glide score as well as the lipophilic efficiency of each compound were calculated simultaneously.
Docking was performed and compared to irfin-1 [18], a previously reported dual inhibitor of the kinase domains of insulin receptor (IR) and insulin growth factor (IGF) . The metabolic pathway of Bacillus subtilis subsp. Subtilis str. OH 131.1 as curated on the Kyoto Encyclopaedia of Genes and Genome (KEGG) [25] was used for pathway enrichment and visualized with Cytoscape 3.5.1 [26].
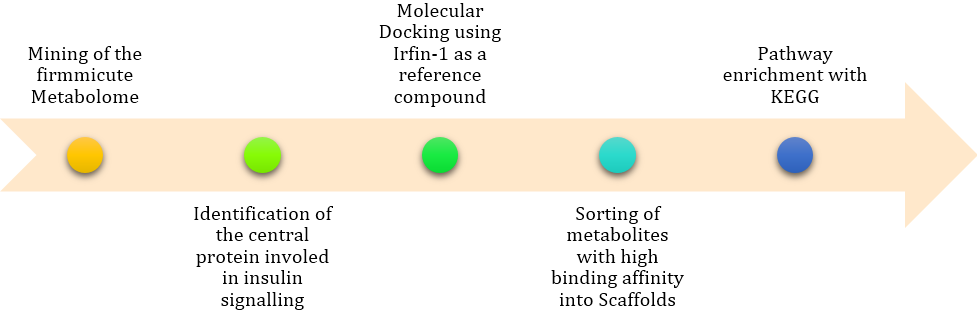
Figure 7: The in-silico pipeline adopted in executing this study. It allowed for the identification of specific pathways that are involved in the inhibition of IRTK.
Conclusion
Despite recent advances in the field of metabolomics and microbiomics, there has been little or no description of how microbial metabolome directly impacts human tissues and cell biology. The data from our study provides for the first time, a direct metabolome-proteome interaction study using an in-silico approach. The public health and economic importance of T2DM in the 21st century makes this strategy a timely one. Nevertheless, the observations are still subject to in vivo validation methods.
References
- Atun R., et al. The Lancet Diabetes & Endocrinology 3.9 (2015): 675-677.
- Zheng Y., et al. Nature Reviews Endocrinology 14 (2017): 88.
- Gonzalez Franquesa A., et al. Current diabetes reports 16.8 (2016): 74.
- Wu T., et al. Journal of proteome research 14.1 (2015): 447-56.
- Aw W., et al. Journal of diabetes investigation 9.1 (2018): 5-12.
- Hossain M U., et al. BioMed research international 2016 (2016): 1-14.
- Segerstolpe A., et al. Cell metabolism 24.4 (2016): 593-607.
- Kadowaki T., et al. Cell 148.3 (2012): 624.
- Samuel V T., et al. Cell 148.3 (2012): 852-871.
- Varshney A., et al. Proceedings of the National Academy of Sciences of the United States of America 114.9 (2017): 2301-2306.
- Catalano K J., et al. PloS one 9.9 (2014): e108693.
- Boucher J., et al. Cold Spring Harbor perspectives in biology 6.1 (2014).
- Napolitano A., et al. PloS one 9.7 (2014): e100778.
- Leal Lopes C., et al. Journal of diabetes research 2015 (2015): 284680.
- Sweeney TE., et al. Best practice & research. Clinical gastroenterology 28.4 (2014): 727-740.
- Zietek T., et al. Frontiers in immunology 7 (2016): 154.
- Palau Rodriguez M., et al. Frontiers in microbiology 6 (2015): 1151.
- Anastassiadis T., et al. Journal of Biological Chemistry 288.39 (2013): 28068-28077.
- Werner H., et al. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology 24.12 (2014): 1947-1953.
- Foster LJ., et al. The Journal of biological chemistry 276.47 (2001): 44212-44221.
- Anastassiadis T., et al. The Journal of biological chemistry 288.39 (2013): 28068-28077.
- Zhao X., et al. Metabolomics 6 (2010): 362-374.
- Suhre K., et al. PloS one 5 (2010): 13953.
- Park J H., et al. Applied Microbiology and Biotechnology 85.3 (2009): 491-506.
- Kanehisa M., et al. Nucleic acids research 44 (2016): 457-462.
- Cline MS., et al. Nature protocols 2.10 (2007): 2366-2382.
Citation:
Adewale J Ogunleye., et al. “Inhibition of Insulin Receptor Tyrosine Kinase by the Firmicutes Metabolome during Insulin
Resistance”. Archives of Endocrinology and Diabetes Care 2.1 (2018): 154-163.
Copyright: © 2018 Adewale J Ogunleye., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.































 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.