Review Article
Volume 1 Issue 2 - 2017
Gene Editing in Animal: Application and Prospect
College of Veterinary Medicine, Haramaya University, PO Box 138 Dire Dawa, Ethiopia
*Corresponding Author: Yimer Muktar, College of Veterinary Medicine, Haramaya University, PO Box 138 Dire Dawa, Ethiopia.
Received: March 03, 2017; Published: June 01, 2017
Abstract
Genetic modification has long provided an approach for reverse genetics i.e. analyzing gene function and linking DNA sequence to
phenotype. However, traditional genome modification technologies have not kept pace with the soaring progress of the genome
sequencing era, due to their inefficiency, time-consuming and labor-intensive features. Recently, invented genome modification technologies,
such asMega nucleases, ZFN (Zinc Finger Nuclease), TALEN (Transcription Activator-Like Effector Nuclease), and CRISPR/
Cas9 nuclease (Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 nuclease) can initiate genome editing easily, precisely
and with no limitations by organism. These new tools have also offered intriguing possibilities in wide range of applications
in animal production sectors including breeding, disease resistance, vector gene manipulation, animal model of human disease and
domestication of animals. Therefore, this review is focused on highlighting the present and possible application of these technologies
along with their practical limitations and socio-economic implication. In conclusion, these technologies are on the raise to address
a wide variety of challenges in animal production involving health aspects, however, due attention must be given to research on the
issues of off-target effects of these technologies, in order to avoid untended modification of animal genomes.
Keywords: Animal; Application; Gene Modification; Method
Introduction
Efficient utilization of genetic engineering has become one of the major challenges of the post genome era. The development of gene targeting in the 1980s has provided the means to perform precise genome manipulation since classical methods prove to be very inefficient, depending on the target locus and the organism [1]. As part of genetic engineering, genome editing is the ability to make specific changes at targeted genomic sites [2].
Gene editing is efficient and precise genetic modification via the induction of a double-strand break in a specific genomic target sequence, followed by the generation of desired modifications during subsequent DNA break repair [3]. Genome sequencing and similar initiatives have increased our understanding of the structure of the genome in many organisms. However, an inability to precisely manipulate any chosen base pair in a genome, particularly in more complex cells and species has meant that functional understandings of genes have lagged behind [4].
The discovery that generating DNA breaks at the target location considerably enhances the efficiency of homologous recombination and paved the way for nuclease-based gene editing. Nucleases are specialized proteins that recognize a specific DNA sequence and cleave it [5]. The first nuclease-based gene editing methods is mega nuclease in which mobile elements created site-specific double-strand breaks by recognizing relatively long sequences i.e. 12-45 bp, and thus have been recognized as an efficient method of gene manipulation ([6] Chevalier and Stoddard, 2001). The second gene editing technology is Zinc Finger Nucleases (ZFNs) in which the nuclease were subsequently created by fusing several linked ‘fingers’ to a sub-part of an endonuclease called the Fok-1 endonuclease. ZFNs recognize 18-36 base-pair DNA sequences [7].
In recent years, targeted gene disruption is becoming a common, facile and essential method for demonstrating the function of a specific gene and is currently performed using transcription activator-like effector nucleases (TALEN) [8] or the clustered regularly interspaced short palindromic repeats (CRISPR/Cas9 system) [9]. These all nuclease based gene editing technologies applied at the embryo level by using micro injection [10].
Gene editing has roles in improvements of milk composition [11,12]. Regarding meat/carcass output, subsequent efforts were made to genetically alter growth rates and patterns to produce of transgenic swine and cattle by targeting their skeletal muscles [13]. In animal breeding, gene editing is considered to remove recessive alleles that impact fertility and increase the frequency of favorable alleles for polygenic traits [14]. Gene editing technologies are also involved in investigation of domestication genes in the domestication process of pigs [15]. Regarding diseases prevention, mammary gland production of antibacterial proteins for enhanced mastitis resistance has been proposed as a prime agricultural application of this technology [16].
As part of the effort to prevent human diseases, animal modeling of human disease is utilized through gene editing technology such as CRISPR-Cas9 in mice as a key ‘model’ organism and monkeys in a disease like lung cancer [17,18]. Another study in pigs as an ideal animal model for studying human cardiovascular disease [19], atherosclerosis [20] and to model infertility and colon cancer using TALENs [21]. On the other hands, gene editing could be applied in vector gene manipulation such as creating genetically modified Anopheles mosquitoes that cannot transmit the malaria pathogens [22]. Overall, this review has the objective of highlighting the role of gene editing technology in improvement of animal production and health including its current application, future prospects and socio ethical implication.
Gene Editing Technology
The principal genome editing techniques are mega nucleases, ZFN, TALEN, and CRISPR. All these techniques achieve precise and efficient genome editing by inducing targeted double-stranded breaks followed by repair. The techniques differ in the way they recognize and break the DNA and in ease of use, efficiency, precision, and cost [23]. Genome editing mediated by ZFNs, TALENs or Cas9 is directly related to the target specificity of these nucleases, since the recognition and cleavage of off-target sites will lead to the introduction of undesired indels (insertion and/or deletion) by non-homologous end joining (NHEJ) repair [24]. If a perfect match between the guide RNA and the target DNA were required forCas9 cleavage, the probability of finding off target by chance would be exceedingly low [25].
The principal genome editing techniques are mega nucleases, ZFN, TALEN, and CRISPR. All these techniques achieve precise and efficient genome editing by inducing targeted double-stranded breaks followed by repair. The techniques differ in the way they recognize and break the DNA and in ease of use, efficiency, precision, and cost [23]. Genome editing mediated by ZFNs, TALENs or Cas9 is directly related to the target specificity of these nucleases, since the recognition and cleavage of off-target sites will lead to the introduction of undesired indels (insertion and/or deletion) by non-homologous end joining (NHEJ) repair [24]. If a perfect match between the guide RNA and the target DNA were required forCas9 cleavage, the probability of finding off target by chance would be exceedingly low [25].
Mega Nuclease System
Mega nucleases are derived from natural nucleases and characterized by long DNA recognition sites allowing them to be used as tools for gene editing; as such long sites are likely to occur. For example, I-SceI, the first gene editing nuclease and the best characterized mega nuclease, recognizes 18 nucleotide-long sequences. The DNA-binding element and the cleaving element are entwined in mega nucleases, which limit the possibility to design new mega nucleases to recognize new targets [5]. It is one of the earliest introduced endonucleases in which mobile elements created site-specific double-strand breaks by recognizing relatively long sequences 12-45 bp, and thus have been recognized as an efficient method of gene manipulation [6].
Mega nucleases are derived from natural nucleases and characterized by long DNA recognition sites allowing them to be used as tools for gene editing; as such long sites are likely to occur. For example, I-SceI, the first gene editing nuclease and the best characterized mega nuclease, recognizes 18 nucleotide-long sequences. The DNA-binding element and the cleaving element are entwined in mega nucleases, which limit the possibility to design new mega nucleases to recognize new targets [5]. It is one of the earliest introduced endonucleases in which mobile elements created site-specific double-strand breaks by recognizing relatively long sequences 12-45 bp, and thus have been recognized as an efficient method of gene manipulation [6].
Zinc Finger Nuclease (ZFN) System
Zinc Finger Nucleases were among the earlier genome editing technologies [26]. It was the first solution developed to overcome mega nuclease limitations in number of potential targets and ease of design. Zinc Finger Nucleases are developed in the 2000s and it is artificial enzymes resulting from the fusion of two protein domains. The DNA-binding element comprises an array of zinc fingers each recognizing a DNA motif. The cleaving element is derived from a nuclease called Fok I. The combination of zinc-finger proteins within a ZFN can be tuned to recognize different target sites, allowing for a greater number of possible targets [5].
Zinc Finger Nucleases were among the earlier genome editing technologies [26]. It was the first solution developed to overcome mega nuclease limitations in number of potential targets and ease of design. Zinc Finger Nucleases are developed in the 2000s and it is artificial enzymes resulting from the fusion of two protein domains. The DNA-binding element comprises an array of zinc fingers each recognizing a DNA motif. The cleaving element is derived from a nuclease called Fok I. The combination of zinc-finger proteins within a ZFN can be tuned to recognize different target sites, allowing for a greater number of possible targets [5].
Existing knowledge about how zinc finger proteins recognize and bind to DNA has now enabled synthetic proteins to be created that incorporate the DNA binding domain of zinc finger proteins. Each zinc ‘finger’ binds to a DNA sequence of 3 base pairs. Zinc Finger Nucleases were subsequently created by fusing several linked ‘fingers’ to a sub-part of Fok-1 endonuclease. ZFNs will recognize 18-36 base-pair DNA sequences [7].
Transcription Activator-Like Effector Nuclease (TALEN) System
Transcription Activator-Like Effector nucleases are artificial enzymes developed from a fusion between a DNA-binding element consisting of an array of TALE subunits and a FokI cleavage element. The TALE subunits, each recognizing a specific DNA nucleotide, independently from the others. This one-to-one correspondence, makes it very easy to design new TALE nucleases and to target any site in the genome [27]. They are more accurate than ZFNs, as the binding DNA sequence is longer ([28] Segal and Meckler, 2013). Researchers have created synthetic TALEs and fused them to the same sub-part of the Fok-1 endonuclease as for ZFNs to create TALENs, which bind to a target sequence of around 13 base pairs [7]. TALE nucleases are both efficient and specific. They exhibit the lowest level of off-target activity when cross-compared with ZFN and CRISPR nuclease [29].
Transcription Activator-Like Effector nucleases are artificial enzymes developed from a fusion between a DNA-binding element consisting of an array of TALE subunits and a FokI cleavage element. The TALE subunits, each recognizing a specific DNA nucleotide, independently from the others. This one-to-one correspondence, makes it very easy to design new TALE nucleases and to target any site in the genome [27]. They are more accurate than ZFNs, as the binding DNA sequence is longer ([28] Segal and Meckler, 2013). Researchers have created synthetic TALEs and fused them to the same sub-part of the Fok-1 endonuclease as for ZFNs to create TALENs, which bind to a target sequence of around 13 base pairs [7]. TALE nucleases are both efficient and specific. They exhibit the lowest level of off-target activity when cross-compared with ZFN and CRISPR nuclease [29].
A key feature of TALENs that distinguishes them from ZFNs is their simple design and assembly parameters. The DNA-binding domain of TALENs consists of tandem repeats of 34 amino acids, with each repeat specifying the binding to a single base pair. TALENs design of any length is simple [30,31]. The repeats within this domain can be reorganized to match a target sequence using routine techniques that are available in most laboratories [8] Cermak., et al. 2011; [32,33].
In addition to simple design and assembly, TALENs have extremely broad targeting range [27], which allows the production of active TALENs for nearly any site within the genome. Using the Goldy TALEN platform, 64% of synthesized TALENs were active in livestock fibroblasts after introduction in to zygotes, with typical activity levels of 20%. Whereas others have failed to produce gene-edited animals by injection of ZFNs into zygotes, they found that several TALEN pairs were efficient at inducing indels by direct injection of TALEN-encoding mRNA into the cytoplasm of both swine and bovine embryos [20].
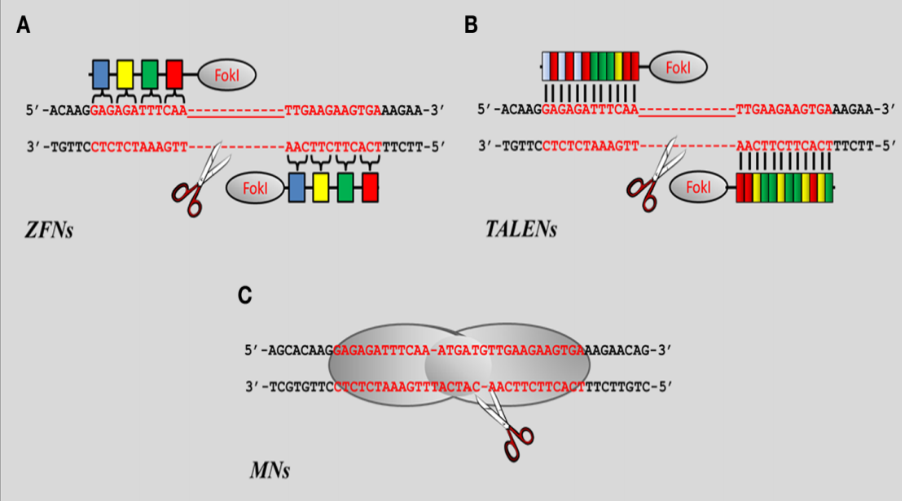
Figure 1: Zink finger nuclease (A), Transcription Activator-Like Effector nuclease
(B) and Mega nuclease (C). Source: Carmen, [34].
(B) and Mega nuclease (C). Source: Carmen, [34].
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 System
Clustered Regularly Interspaced Short Palindromic Repeats or Cas9 genome editing comprises a Cas9 endonuclease guided to its target sequence by a specifically designed guide RNA of around 20 base pairs. It is a naturally occurring system in prokaryotic cells [4]. However, it works in both simple and more complex cells [35]. Recent fast and massive adoption of this technique is due to its simplicity, speed, and low cost to design a new CRISPR nuclease requiring only a new guide RNA sequence with a high efficiency [36].
Clustered Regularly Interspaced Short Palindromic Repeats or Cas9 genome editing comprises a Cas9 endonuclease guided to its target sequence by a specifically designed guide RNA of around 20 base pairs. It is a naturally occurring system in prokaryotic cells [4]. However, it works in both simple and more complex cells [35]. Recent fast and massive adoption of this technique is due to its simplicity, speed, and low cost to design a new CRISPR nuclease requiring only a new guide RNA sequence with a high efficiency [36].
CRISPR Mechanism: There are three steps to undertake gene editing through CRISPR. The first step is Spacer Integration, in which new spacers are inserted at the leader end of the CRISPR array when DNAs like bacterial DNA are infected with foreign DNA [37]. However, this mechanism does not cause CRISPRs to expanding definitely; when a new spacer is inserted, a different spacer is generally deleted. New spacer integration allows for organisms to adapt immunity based on the variety of phages present in their current environment, while retaining relevant ancestral spacers [38].
The second step is crRNA Transcription; Cas genes encode four Cas proteins, including Cas1 and Cas2 proteins which are universal in all CRISPR systems. These systems also express Cas4 which is involved in spacer integration. Finally, all CRISPR systems encode a highly conserved Cas9 gene [39]. Long primary pre-crRNA is transcribed from the CRISPR loci. Pre pairs (pre-crRNA) with trans-activating CRISPR RNA (tracrRNA) will be processed by RNase III [37]. This creates shorter mature crRNAs. Then Cas9 catalyzes the formation of mature crRNA-tracrRNA complex from shorter mature crRNAs [39].
The third step, CRISPR Gene Editing is followed after transcription, processing and mature crRNAs- tracrRNA complex formation with Cas9. This complex binds to a protospacer sequence of extra-chromosomal double-stranded DNA. The process is dependent on a protospacer adjacent motif (PAM) [39]. When the Cas9/tracrRNA/crRNA complex binds the target sequence of the dsDNA, an R-loop forms and one DNA strand with crRNA and the other disassociates. Both strands of DNA are cut near the PAM sequence. The crRNA acts as a guide while Cas9 acts as the endonuclease to cleave the DNA. The presence of double stranded breaks (DSB) in the DNA leads to activation of the DSB repair machinery either Non-homologous end joining (NHEJ) or homology-directed repair(HDR)as indicated in the bellow figure (figure 1) [35].
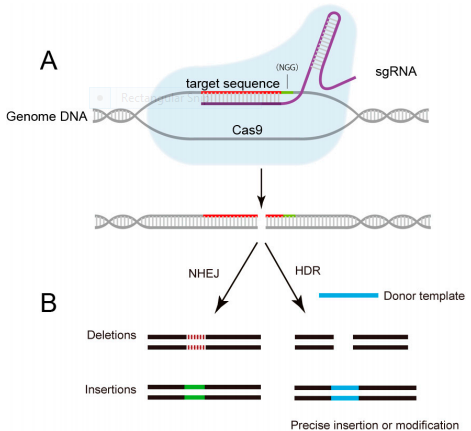
Figure 2: CRISPR/Cas9 mediated-genome editing Lin., et al. [40].
NGG (nucleobase followed by two guanine).
CRISPR system includes Cas proteins, gene scissors that can bind specific RNA sequences, which subsequently act as guide molecules. The bound guide RNA ultimately determines which complementary DNA within the genome is targeted and precisely cut by the gene scissors [9]. The first advantage is the greater ease by which CRISPR/Cas can be designed and implemented. Unlike ZFNs and TALENs which require the complex design of zinc-finger and TALE DNA binding arrays for every new genomic target site, CRISPR/Cas simply requires changing the 20-nt sgRNA spacer sequence so that it matches the target site [29]. The second advantage of CRISPR/Cas is its comparable or greater genome-editing efficiencies than that of ZFNs or TALENs. In general, CRISPR/Cas functions with greater consistency, efficacy, and less toxicity than lab-produced ZFNs [41], and it is likely to be more effective at targeting methylated genomic sites compared to TALENs [42].
| Mega Nuclease | ZFN | TALEN | Cas9 | |
| Length of Target Site | 12-45 bp | 18-36bp | 30-40 bp | 22 bp |
| Off-Target Effects | Low | Variable | Low | Variable |
| Nuclease Size | 1 kb | 1 kb | 3 kb | 4.2 kb |
| Accessibility for Engineering | Difficult | Difficult | Moderate | Easy |
| Multiple Targeting | Difficult | Difficult | Difficult | Easy |
| In vivo Delivery | Easy | Easy | Difficult | Moderate |
| DNA Targeting System | Protein | Protein | Protein | Protein/RNA |
Table 1: Comparison of Different Nuclease Systems. Tsai [43].
Gene Editing Applications
Animal Breeding and Production
The ability to generate edited gene is an extremely powerful tool for the analysis of gene function and for the generation of animals with biotechnological or breeding applications [44]. From an animal breeding perspective, there are three immediately obvious roles for genome editing: fix favorable alleles for monogenic traits such as disease resistance, myostatin, polledness. Secondly, remove recessive alleles that impact fertility and finally to increase the frequency of favorable alleles for polygenic traits [45].
Animal Breeding and Production
The ability to generate edited gene is an extremely powerful tool for the analysis of gene function and for the generation of animals with biotechnological or breeding applications [44]. From an animal breeding perspective, there are three immediately obvious roles for genome editing: fix favorable alleles for monogenic traits such as disease resistance, myostatin, polledness. Secondly, remove recessive alleles that impact fertility and finally to increase the frequency of favorable alleles for polygenic traits [45].
Meat Production: Myostatin is a member of the transforming growth factor beta (TGF-β) super family, involved in the inhibition of muscle differentiation and growth [46] and the inhibition of this protein induces a significant increase in the muscle volume and mass producing more meat in animals known as double-muscle animals [47]. Myostatin have been previously modified by genetic engineering in ruminant species using technologies such as ZFN and TALEN. However, the use of the CRISPR/Cas9 system is easier and is now available for livestock production [48].
In study on disruption of genes in goat primary fibroblasts by CRISPR/Cas9-mediated approach, only the myostatin (MSTN) knockout fibroblasts were achieved that targets the skeletal muscle [49]. ZFN cleavage-triggered NHEJ yielded biallelic modifications of the bovine myostatin gene in which Myostatin protein levels in somatic cell nuclear transfer (SCNT) produced calves were reduced by about 50% and the animals displayed a double muscled phenotype at the age of one month [50].
Milk Production: Genome editing technology was used to improve the composition of cows’ milk by preventing the expression of the allergenic milk protein beta lacto globulin (BLG). NHEJ was harnessed to modify the bovine gene for BLG following ZFN cleavage near the start codon of the gene. Cells with heterozygous biallelic modifications were used as donors for the generation of live offspring. However, the two allelic variants present in the surviving calf had small in-frame deletions which did not disrupt the BLG reading frame and thus modified the gene ([51] Yu., et al. 2011). Milk proteins are coded by unique copy genes that can be altered to modify milk composition and properties. Different applications of milk modification in transgenic animals have various roles in improvements of milk composition [11,12].
Firstly, to modify bovine milk to make it more appropriate to the consumption of infants. Human milk has a high content of lactoferrin and lysozyme when compared to bovine milk. Lactoferrin is responsible for the iron transport and inhibits the bacterial growth. Introduction of human lactoferrin into the bovine milk and elimination of the β-lacto globulin from the cow milk since it is the major allergens of cow's milk have been achieved through genetic engineering [52]. Secondly, to reduce the content of lactose in the milk to allow its consumption to people with intolerance to lactose. It is considered that 70% of the world population is lacking the intestinal lactase, the enzyme required to digest the lactose. The reduction in lactose may be obtained by expressing β-galactosidase in the milk or diminishing the content of α-lactalbumin [53].
On the other hands, to alter the content of caseins of the milk to increase its nutritive value, cheese yield and processing properties. Research has intended to increase the number of copies of the gene of the κ-casein, to reduce the size of the micelles and modifying the κ-casein to make it more susceptible to the digestion with chymosin [54]. Modified female bovine foetal fibroblasts to express additional copies of transgenes encoding two types of casein: bovine β-casein and κ-casein. The modified cell lines of fibroblasts were used to create eleven cloned calves. Milk from the cloned animals was enriched for β- and κ-casein, resulting in 30% increase in the total milk casein and 13% increase in total milk protein, demonstrating the potential of this technology to produce modified milk [55]. Finally, gene editing technologies can also be used to express antibacterial substances in the milk, such as proteases, lysozyme or transferrin in the milk to increase concentrations of antibacterial proteins and mastitis resistance [56].
Wool Production: Beside the focus on meat, additional purposes in sheep were increased wool production and improved fiber with better processing and wearing qualities by ZFN ([57] Bawden., et al. 1998). The quality, color, yield and ease of harvest of hair, wool and fiber for fabric and yarn production have been an area of focus in livestock production. The manipulation of the quality, length, fineness and crimp of wool, hair and fiber from sheep and goats has been examined using transgenic methods [58].
The generation of genetically modified animals by using CRISPR system has several advantages for different applications not only in meat, dairy but also in wool production [59]. The objectives are to improve production of sheep wool and to modify the properties of the fibbers and since cysteine seems to be the limiting amino acid for wool synthesis, the first approach was to increase its production through transfer of cysteine biosynthesis from bacterial genes to sheep genome [12].
Domestication of Animals
Domestication is an evolutionary process driven by artificial selection, and the underlying mechanisms are still unclear. By comparing the genomes of domesticated and wild species, numerous candidate genes are predicted to be involved in the domestication process. For example, there are about 516 reported candidate domestication genes for pigs and 354 domestication genes for silkworm [15,60].
Domestication is an evolutionary process driven by artificial selection, and the underlying mechanisms are still unclear. By comparing the genomes of domesticated and wild species, numerous candidate genes are predicted to be involved in the domestication process. For example, there are about 516 reported candidate domestication genes for pigs and 354 domestication genes for silkworm [15,60].
Traditional genetic modification technology cannot handle large numbers of genes in a short period of time, TALEN and CRISPR/Cas9 methods may provide a bridge to overcome this challenge and mediate the epigenetic status of genes [61]. Additionally, two or more genes could be modified at the same time using these technologies [62]. Domesticated species genomes have beenmodified or are undergoing experiments through TALEN and CRISPR/Cas9 in pig [23,63], goat and cattle [21].
Disease Resistance
Mastitis costs the dairy industry billions of dollars annually and is the most consequential disease of dairy cattle. Mammary gland production of antibacterial proteins for enhanced mastitis resistance has been proposed as a prime agricultural application of gene editing technology [16]. Consequently, transgenic cows secreting an antimicrobial peptide demonstrated resistance to mastitis. Recent studies found that a precisely placed double strand break induced by engineered ZFNs stimulated the integration of exogenous DNA stretches into a pre-determined genomic location, resulting in high-efficiency site-specific gene addition. ZFNs was used to target human lysozyme (hLYZ) gene to bovine b-casein locus, resulting in hLYZ knock-in of approximately 1% of ZFN-treated bovine fetal fibroblasts (BFFs). The production of cloned cows carrying human lysozyme gene knock-in b-casein locus using ZFNs is for the creation of transgenic cows by genetic engineering to provide a viable tool for enhancement of not only mastitis resistance but also prevention of other pathogens and improve health [56].
Mastitis costs the dairy industry billions of dollars annually and is the most consequential disease of dairy cattle. Mammary gland production of antibacterial proteins for enhanced mastitis resistance has been proposed as a prime agricultural application of gene editing technology [16]. Consequently, transgenic cows secreting an antimicrobial peptide demonstrated resistance to mastitis. Recent studies found that a precisely placed double strand break induced by engineered ZFNs stimulated the integration of exogenous DNA stretches into a pre-determined genomic location, resulting in high-efficiency site-specific gene addition. ZFNs was used to target human lysozyme (hLYZ) gene to bovine b-casein locus, resulting in hLYZ knock-in of approximately 1% of ZFN-treated bovine fetal fibroblasts (BFFs). The production of cloned cows carrying human lysozyme gene knock-in b-casein locus using ZFNs is for the creation of transgenic cows by genetic engineering to provide a viable tool for enhancement of not only mastitis resistance but also prevention of other pathogens and improve health [56].
On a separate effort, porcine reproductive and respiratory syndrome is the most economically important disease of swine caused by the porcine reproductive and respiratory syndrome virus. CRISPR-Cas9 has been utilized to generate pig that are resistant to the porcine reproductive and respiratory syndrome virus [64].
Animal Model of Human Disease
CRISPR-Cas9 is being used in mice; a key ‘model’ organism and the birth of transgenic monkeys engineered using this system has also recently been reported [17,18]. Pig is often considered as an ideal animal model for studying human cardiovascular disease [19], atherosclerosis [20] and to model infertility and colon cancer using TALENs [21]. TALENS also has been used in frogs, rats, and cows; among others for modelling human diseases [29]. CRISPR-Cas9-mediated genome editing of tumor suppressor genes has been shown to enable the rapid functional characterization of putative lung cancer genes using mouse models [65]. Zebra fish is also a powerful and tractable animal model for functional genomics analysis and investigating of human disease pathogenesis, as well as, for the discovery and development of new drugs [66].
CRISPR-Cas9 is being used in mice; a key ‘model’ organism and the birth of transgenic monkeys engineered using this system has also recently been reported [17,18]. Pig is often considered as an ideal animal model for studying human cardiovascular disease [19], atherosclerosis [20] and to model infertility and colon cancer using TALENs [21]. TALENS also has been used in frogs, rats, and cows; among others for modelling human diseases [29]. CRISPR-Cas9-mediated genome editing of tumor suppressor genes has been shown to enable the rapid functional characterization of putative lung cancer genes using mouse models [65]. Zebra fish is also a powerful and tractable animal model for functional genomics analysis and investigating of human disease pathogenesis, as well as, for the discovery and development of new drugs [66].
Mouse models has been used as cystic fibrosis modeling, however, it does not fully recapitulate the natural progression of cystic fibrosis of lung and pancreatic disease seen in human patients [67]. For these reasons, two additional cystic fibrosis models were generated i.e. pig and ferret. Findings from these larger cystic fibrosis animal models have begun to demonstrate many phenotypic similarities to the human disease [68]. It is possible that such animal models of neurodegenerative disease can be used to develop effective treatment for different protein misfolding diseases and model of Alzheimer’s disease [69,70].
Vector Gene Manipulation
Gene editing by using gene drive to introduce modified genes into wild-type pest populations in a bid to render them harmless [71]. It could be applied, for example, in the fight against malaria and dengue fever or in the containment of invasive species, such as cane toads and rats. Malaria pathogens are transmitted to humans by the Anopheles mosquito. But, it is possible to reduce transmission by creating genetically modified Anopheles mosquitoes that cannot transmit the malaria pathogens. Further development in fruit fly models involves a genetic element based on the CRISPR-Cas9 system being inserted into the genome of the fly [22]. TALEN-based editing was also associated with small deletions ranging in size from 1-7 bp. This is mostly used in mosquitoes and similar results have been obtained in other insects such as vinegar fly D. melanogaster [72], the silkworm B. mori [73] and the cricket G. bimaculatus [74].
Gene editing by using gene drive to introduce modified genes into wild-type pest populations in a bid to render them harmless [71]. It could be applied, for example, in the fight against malaria and dengue fever or in the containment of invasive species, such as cane toads and rats. Malaria pathogens are transmitted to humans by the Anopheles mosquito. But, it is possible to reduce transmission by creating genetically modified Anopheles mosquitoes that cannot transmit the malaria pathogens. Further development in fruit fly models involves a genetic element based on the CRISPR-Cas9 system being inserted into the genome of the fly [22]. TALEN-based editing was also associated with small deletions ranging in size from 1-7 bp. This is mostly used in mosquitoes and similar results have been obtained in other insects such as vinegar fly D. melanogaster [72], the silkworm B. mori [73] and the cricket G. bimaculatus [74].
Limitation and Prospects of Gene editing Technologies
TALEN and CRISPR/Cas9 systems are promising accurate genome editing tools that have the potential to promote biological research. However, there are limitations to both techniques. With regards to TALEN, the plasmid is large, which would affect delivery efficiency to cells, and it is difficult to assemble repeat monomers while the main weakness of CRISPR/Cas9 is the occasional high off-target effects, in special species or gene cases [75]. Efforts could be made to minimize the limitation of both technologies. For TALENs, developing a new TALEN scaffold would diminish the plasmid size, and different kits have been invented enabling monomer assembly in a short time. For CRISPR/Cas9, a pair of Cas9 nicking variants that requires cooperatives to generate a DSB would reduce the likelihood of off-target effects [76].
TALEN and CRISPR/Cas9 systems are promising accurate genome editing tools that have the potential to promote biological research. However, there are limitations to both techniques. With regards to TALEN, the plasmid is large, which would affect delivery efficiency to cells, and it is difficult to assemble repeat monomers while the main weakness of CRISPR/Cas9 is the occasional high off-target effects, in special species or gene cases [75]. Efforts could be made to minimize the limitation of both technologies. For TALENs, developing a new TALEN scaffold would diminish the plasmid size, and different kits have been invented enabling monomer assembly in a short time. For CRISPR/Cas9, a pair of Cas9 nicking variants that requires cooperatives to generate a DSB would reduce the likelihood of off-target effects [76].
Cas9 protein and FokI protein have been combined to forma dimeric CRISPR/Cas9 RNA-guided FokI nucleases system, which could be useful in highly accurate genome editing applications [77]. A strict screening strategy of seed region on the region of sgRNA would help decrease un-desired mutagenesis in an off-target region. Studies have shown that seed region accounts for most of CRISPR/Cas9specificity [9] and a point mutation in this region would abrogate sgRNA: Cas9recognition [25]. Hence, at least two mismatches of a seed region lying in the off-target sequence would improve targeting specificity complimentary to a target region would dramatically reduce the off-target effects by 5000 fold, without scarifying target efficiency [78].
Generally, Cas9-based technologies are spreading at a remarkable speed that speaks to their simplicity and efficiency. One critical point is the precise determination of off-target effects and their consequences. Although current data seem to indicate that these are not a major obstacle, a better characterization of Cas9 cleavage is required before the technology is used in animals [79].
Socio Economic Importance
Genome edition leading to greater production of transgenic animals have medical benefits such as potential to use animals to develop pharmaceuticals and vaccines, thereby eliminating risks and maximizing benefits to humans, and potential to have healthier animals, free from disease or better suited to certain environments [80]. Questions of social justice for the farming industry such as small scale farmers being negatively impacted by the market dominance of those able to afford genetically modified animals, or the dominance of those companies able to utilize genetic modification technology and breed livestock from it. There is also the potential for the loss of traditional farming practices should farming using genetically modified animals become the norm [81]. Although, patent protection might prevent or hinder introduction of genetically modified livestock in developing nations, there is potential for vast gains in terms of quality, supply of livestock and associated product for human use [80].
Genome edition leading to greater production of transgenic animals have medical benefits such as potential to use animals to develop pharmaceuticals and vaccines, thereby eliminating risks and maximizing benefits to humans, and potential to have healthier animals, free from disease or better suited to certain environments [80]. Questions of social justice for the farming industry such as small scale farmers being negatively impacted by the market dominance of those able to afford genetically modified animals, or the dominance of those companies able to utilize genetic modification technology and breed livestock from it. There is also the potential for the loss of traditional farming practices should farming using genetically modified animals become the norm [81]. Although, patent protection might prevent or hinder introduction of genetically modified livestock in developing nations, there is potential for vast gains in terms of quality, supply of livestock and associated product for human use [80].
Conclusions and Recommendations
Even though the utilization of gene editing is still in its early stages and there are various concerns including ethical, socio-economical aspect since unregulated alteration of the natural genetic balance may lead into undesirable consequences due to off target gene edition. There is a reason to believe that it will make the exploration of gene functions more precise and in depth in the near future. From animal production perspective, empowered by these technologies animal health professionals will now be able to utilize these technologies in various areas such as animal production, animal modeling, disease resistance, vector gene manipulation, animal breeding, and domestication by using artificially synthesized nucleases since the previous technologies uses genes from other animals. In addition, given the inexpensiveness of these technologies, they are potentially useful and applicable even in developing countries. Based on the above conclusion, intensive research should be employed in order to alleviate some of the draw backs of these recent technologies such as the off-target effects. In addition, continuous and rigorous effort should be done to further utilize the potential of these technologies for enhancement of livestock productivity and ensuring food security along with minimizing the socio-economic implication of gene editing.
Conflict of Interests
The authors declared no conflict of interests regarding the present paper.
The authors declared no conflict of interests regarding the present paper.
Authors’ Contribution
NT conceived the idea DA and YM, collected and studied published papers, drafted the review paper, all participated in conceptualization of the idea, study design, review, and editing the paper.
NT conceived the idea DA and YM, collected and studied published papers, drafted the review paper, all participated in conceptualization of the idea, study design, review, and editing the paper.
References
- Smithies. “Forty years with homologous recombination”. Nature Medicine 7 (2001): 1083-1086.
- AJ Bogdanove and DF Voytas. “TAL Effectors Customizable proteins for DNA targeting”. Science 333.6051 (2011): 1843-1846.
- YG Kim., et al. “Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain”. Proceedings of the National Academy of Sciences 93.3 (1996): 1156-1160.
- CA Gersbach “Genome engineering: the next genomic revolution”. Nature Methods 11.10 (2014): 1009-1011.
- B Elsy and R Magali. “Rewriting the Book of Life: New Era in Precision Gene Editing”. Bcg Perspectives 13. (2015).
- BS Chevalier and BL Stoddard. “Homing endonucleases: strucrural and functional insights in to the catalysis of intro/intein mobility”. Nucleic acids Research 29.18 (2001): 3757-3774.
- N De Souza. “Primer Genome editing with engineered nucleases”. Nature Methods 9.1 (2012): 27.
- T Cermak., et al. “Efficient design and assembly of custom TALEN and other TAL effector-based constructs forDNA targeting”. Nucleic Acids Research 39.12 (2011): 82.
- M Jinek., et al. “A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity”. Science 337. 6096 (2012): 816-821.
- W Qin., et al. “Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease”. Genetics 200.2 (2015): 423-430.
- EA Maga and JD Murray. “Mammary gland expression of transgenes and the potential for altering the properties of milk”. Nature Biotechnology 13.13 (1995): 1452-1457.
- JD Murray., et al. “Transgenic animals in Agriculture”. Nature 409. 6822 (2001): 860-921.
- ER Cameron., et al. “Transgenic science”. The British Veterinary Journal 150.1 (1994): 9-24.
- Y Jiang., et al. “Multi gene editing in the Escherichia coligenome using the CRISPR-Cas9 system”. Applied Environmental Microbiology 81.7 (2015): 2506-2514.
- M Li., et al. “Genome editing in large animal”. Giga Science 3.24 (2014): 9-10.
- RJ Wall., et al. “Genetically enhanced cows resist intramammary Staphylococcus aureus infection”. Nateture Biotechnology 23.4 (2005): 445-451.
- DW Harms., et al. “Mouse Genome Editing Using the CRISPR/Cas System”. Current Protocols in Human Genetics 15.7 (2014): 15-27.
- Y Niu., et al. “Generation of gene-modified cynomolgus monkey via Cas9/RNA- mediated gene targeting in one-cell embryos.” Cell 156.4 (2014): 836-843.
- L Yang., et al. “RNA Guided Human Genome Engineering via Cas9”. Science 339. 6121 (2013): 823-826.
- DF Carlson., et al. “Efficient TALEN-mediated gene knockout in livestock”. Proceedings of the National Academy of Sciences 109.43 (2012): 17382-17387.
- WF Tan., et al. “Efficient nonmeiotic allele introgression in livestock using custom endonucleases”. Proceedings of the National Academy of Sciences 110.41 (2013): 16526-16531.
- CF Gantz and E Bier. “The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations”. Science 348.6233 (2015): 442-444.
- L Gasiunas and P Jinek. “Gene editing mediated by ZFN, TALEN orCas9 system”. Science 337. (2012): 816-821.
- E Deltcheva., et al. “CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III”. Nature 471.7340 (2011): 602-607. 2011.
- WY Jiang., et al. “RNA-guided editing ofbacterial genomes using CRISPR-Cas systems”. Nature Biotechnology 31.3 (2013): 233-239.
- FD Urnov., et al. “Genome editing with engineered zinc finger nucleases”. Nature Reviews Genetics 11.9 (2010): 636-46.
- L Cade., et al. “Highly efficient generation of heritable zebra fish gene mutations using homoand heterodimeric TALENs”. Nucleic Acids Resareach 40.16 (2012): 8001-8010.
- DJ Segal and JF Meckler. “Genome establishment engineering at the dawn of the golden age”. Nature 14. (2013): 135-158.
- JK Joung and JD Sander. “TALENs: A widely applicable technology for targeted genome editing”. Nature Reviews Molecular Cell Biology 14.1 (2013): 49-55.
- J Boch., et al. “Breaking the code ofDNA binding specificity of TAL-TypeIIIeffectors”. Science 326.5959 (2009): 1509-1512.
- MJ Moscou., et al. “A simple cipher governs DNA recognition by TAL effectors”. Science 326.5959 (2009): 1501.
- T Li., et al. “Modularly assembled designer TALeffector nucleases for targeted gene knockout and gene replacement in eukaryotes”. Nucleic Acids Research 39.14 (2011): 6315–6325.
- R Morbitzer., et al. “Assembly of custom TALE-type DNA binding domains by modular cloning”. Nucleic Acids Research 39.13 (2011): 5790–5799.
- B Carmen. “Emerging gene editing strategies for Duchenne muscular dystrophy targeting stem cells. Review”. Frontiers in Physiology 148. 5 (2014) 1-7.
- P Mali., et al. “Church Cas9 as a versatile tool for engineering biology”. Nature methods 10.10 (2013): 957-963.
- CM Yasue and Pankaj. “Cell Stem Cell”. Scientific Reports 4. (2014): 331-335.
- LA Marraffini and EJ Sontheimer. “CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea”. Nature Reviews Genetics 11.3 (2010) 181-190.
- R Barrangou., et al. “CRISPR provides acquired resistance against viruses’ in prokaryotes”. Science 315.5819 (2007): 1709-1712.
- L Cong., et al. “Multiplex genome engineering using CRISPR/Cas systems”. Science 339.6121 (2013): 819–823.
- G Lin and JL Kuozhang, “Genetic modification through ZFNs”. International Journal of Molecular Sciences 16. (2015): 2607-26086.
- TI Cornu., et al. “DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases”. Molecular Therapy 16.2 (2008): 2352-358.
- PD Hsu., et al. “DNA targeting specificity of RNA-guided Cas9 nucleases”. Nature Biotechnology 31.9 (2013): 827–832.
- SQ Tsai., et al. “GUIDE-seq enablesgenome-wideprofilingof off-targetcleavagebyCRISPR-Casnucleases”.Nature Biotechnology 33.2 (2015): 187–197.
- SC Fahrenkrug., et al. “Glenn, Precision genetics for complex objectives in animal agriculture”. Journal of Animal Science 88.7 (2010): 2530–2539.
- J Fritch. “Opportunities and limits of gene editing”. Nature science 4. (2015): 22-24.
- SJ Lee. “Regulation of muscle mass by myostatin”. Annual Review of Cell and Developmental Biology 20. (2004): 61–86.
- BNK Le.,et al. “Myostatin inhibition enhances the effects of exercise on performance and metabolic outcomes in aged mice”. Journal Gerontology A Biology Science Medical Science 64.9 (2009): 940–948.
- C Proudfoot., et al. “Genome edited sheep and cattle”. Transbondary Research 24.1 (2014): 147–53.
- W Ni. “Efficient gene knockout in goats using CRISPR/Cas9 system”. Cell 9.9 (2014): 1061.
- J Luo., et al. “Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases”. PloS One 9.4 (2014): e95225.
- S Yu., et al. “Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle”. Cell Research 21.11 (2011) 1638–1640.
- B Van HC Patrick., et al. “Large scale production of recombinant human lactoferrin in the milk of transgenic cows”. Nature Biotechnology 20.5 (2002): 484-487.
- MG Stinnakre., et al. “Creation and phenotypic analysis of alpha-lactalbumin-deficient mice”. Proceedings of the National Academy of Sciences 91.14 (1994): 6544-6548.
- A Gutierrez-Adan., et al. “Alterations of the physical characteristics of milk from transgenic mice producing bovine κ-Casein”. Journal of Dairy Science 79.5 (1996): 791-799.
- B Brophy., et al. “Cloned transgenic cattle produce milk with higher levels of beta-casein and kappa-casein”. Nature Biotechnology 21.2 (2003): 157-162.
- X Liu., et al. “Generation of mastitis resistance in cows by targeting human lysozyme gene to b-casein locus using zinc-finger nucleases”. Proceedings. Biological Sciences 281.1780 (2014): 1-10.
- CS Bawden., et al. “Expression of a wool intermediate filament keratin transgene in sheep fibre alters structure”. Transgenic Research 7.4 (1998): 273–287.
- G Scott and B Mattew. Genetically engineered animals and public health: Compelling Benefits for Health Care, Nutrition, and the Environment, and Animal Welfare. (2011): 22-30.
- H Han., et al. “One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system”. Frontiers of Environmental Science and Engineering 1.1 (2014): 2-5.
- J Jensen., et al. “Complete resequencing of 40 genomes reveals domesticationevents and genes in silkworm”. Science 326. 5951 (2009): 433-436.
- ML Maeder., et al. “Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins”. Nature Biotechnology 31.12 (2013): 1137-1142.
- H Wang., et al. “One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering,” Cell 153.4 (2013): 910-918.
- SG Lillico., et al. “Live pigs produced from genome edited zygotes”. Scientific Reports - Nature 3. (2013): 2847.
- W Whit., et al. “Gene edited pigs are protected from porcine reproductive and respiratory syndrome virus”. Nature Biotechnology 34.1 (2016): 20-22.
- FJ Sanchez., et al. “Rapid modeling of cooperating genetic events in cancer through somatic genome editing”. Nature 516.7531 (2014): 428-431.
- H Campbell., et al. “Highly efficient generation of heritable zebra fish gene mutations using homoand heterodimeric, TALENs”. Nucleic Acids Research 40.16 (2012): 8001–8010.
- JT Fisher., et al. “Comparative biology of cystic fibrosis animal models”. Methods in Molecular Biology 742. (2011): 311–334.
- X Sun., et al. “Disease phenotype of a ferret cftr-knockout model of cystic fibrosis”. Journal of Clinical Investigation 120.9 (2010): 3149–3160.
- R Crook., et al. “A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin1”. Nature Medicine 4.4 (1998): 452–455.
- R Chang., et al. “Transgenic animal models for study of the pathogenesis of Huntington’s disease and therapy”. Drug Design, Development and Therapy 9. (2015): 2179–2188.
- CKA Oye., et al. “Regulating gene drives”. Science 345.6197 (2014): 626-628.
- J Liu., et al. “Efficient and specificmodifications of the Drosophila genome by means of an easy TALEN strategy”. Journal of Genetics and Genomics 39.5 209-215.
- S Sajwan., et al. “Efficientdisruption of endogenous Bombyx gene by TAL effector nucleases”. Insect Biochemistry and Molecular Biology 43.1 (2013): 17-23.
- T Watanabe., et al. “Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases”. Nature Communications 3. (2012): 1017.
- V Pattanayak., et al. “High through put profiling of off-target DNA cleavage reveals RNA-programmed Cas9nuclease specificity”. Nature Biotechnology 31.9 (2013): 839–843.
- FA Ran., et al. “Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity”. Cell 154.6 (2013): 1380–1389.
- SQ Tsai., et al. “Dimeric CRISPR RNA-guided FokI nucleases for highlyspecific genome editing”. Natuer Biotechnololgy 32.6 (2014): 569-576.
- YF Fu., et al. “Improving CRISPR-Casnuclease specificity using truncated guide RNAs”. Nature Biotechnology 32.3 (2014): 279–284.
- E Pennisi. “The CRISPR craze”. Science 341.6148 (2013): 833–836.
- RD Combes and M Balls. “Every silver lining has a cloud: the scientific and animal welfare issues surrounding a new approach to the production of transgenic animals”. Alternatives to Laboratory Animals 42.2 (2014): 137-145.
- J Kimmelman. “The ethics of human gene transfer”. Nature Reviews Genetics 9.3 (2008): 239-44.
Citation:
Yimer Muktar., et al. “Gene Editing in Animal: Application and Prospect”. Multidisciplinary Advances in Veterinary Science 1.2
(2017): 57-67.
Copyright: © 2017 Yimer Muktar., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.





























 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.