Research Article
Volume 2 Issue 1 - 2017
Candida albicans RAPD-PCR Genotypic Study in Peri-implant Sulcus and Buccal Mucosa
1University of Buenos Aires, Argentina. School of Medicine. Department of Microbiology, Parasitology and Immunology
2University of Buenos Aires, Argentina School of Dentistry Department of Microbiology and Parasitology
2University of Buenos Aires, Argentina School of Dentistry Department of Microbiology and Parasitology
*Corresponding Author: Virginia M Jewtuchowicz, University of Buenos Aires, Argentina. School of Medicine. Department of Microbiology, Parasitology and Immunology.
Received: November 24, 2017; Published: November 28, 2017
Abstract
Molecular techniques have been used in recent studies to identify a wide range of potential bacterial pathogens in periimplant pockets of the oral cavity. However, the prevalence and molecular epidemiology of yeasts and species distribution related to periimplantitis are as yet unknown. The aim of this study was to determine the prevalence and distribution of yeasts in periimplant biofilm and to study genetic relatedness of Candida albicans. Yeasts recovered from periimplant biofilm samples (n = 242) and buccal samples (n = 300) were studied in 100 immunocompetent nonsmoking patients who visited the dental clinic of the Asociación Implantodontológica Argentina, Buenos Aires, Argentina, and had received oral rehabilitation with implants for more than five years. Yeasts recovered from samples were studied by typing assays using RAPDPCR. The prevalence of yeasts in the periimplant sulcus was 37.6% (n = 91/242, CI 95%: 31.5 43.7). C. albicans was the most prevalent species identified in this study population. The RAPD analysis showed identical genotypes in most C. albicans spp. from the two different sampling sites: buccal and periimplant. These findings suggest that periimplant biofilm is an ecological niche that favors the growth of yeast species. Most C. albicans found in periimplant biofilm originate from the endogenous infection caused by commensal strains.
Keywords: Implants; biofilm; Candida albicans; RAPDPCR; Periimplantitis
Introduction
The use of osseointegrated implants, as well as their complications and problems, have increased inrecent decades. Successfully osseointegrated titanium implants usually harbor low quantities of plaque and present little marginal inflammation. Supraand subgingival microbiota at well maintained implant sites seem to resemble the microbiota associated with healthy gingiva. An increased proportion of putative periodontal pathogens has been documented at implant sites suggesting that the periodontal pocket may serve asa reservoir for colonization of titanium implants. Periimplantitis is a chronic progressive marginal infection, defined as an inflammatory reaction that affects the tissue surrounding osseointegrated dental implants, resulting in the loss of the supporting bone. Microbiota resembling that of adult peiodontitis has been found in periimplantitis [1-4].
Extensive antibiotic treatment and irrigation with chlorhexidine may cause etiological changes. Microorganisms not primarily associated with periodontitis, such as Staphylococcus spp., enterics and Candida spp., have also been isolated [2-5]. Molecular techniques have been used in recent studies to identify a wide range of potential bacterial pathogens in periimplant pockets [6,7]. However, the prevalence of yeasts and species distribution related to periimplantitis are as yet unknown. The same has been found to be true for dental biofilm [2,8]. Dahlen., et al.[9] and Reynaud., et al.[1] claim that there was colonization by the genus Candida spp. in periodontal pockets, refractoryperiodontitis [3,10,11], and implant failure. Other studiesreport presence of Candida albicansin thesubgingival plaque microbiota of human immuno deficiencyvirus (HIV) positive individuals [12]. In recent years, several molecular typing methodshave been used to characterize Candida spp. Isolates and to delineate strain relatedness, the mostwidely used being polymerase chain reaction (PCR) based methods.
Among these, the random amplifiedpolymorphic DNA (RAPD) method of DNAfingerprinting has become quite popular for allinfectious fungi and has been successfully appliedto assess the genetic relatedness of Candida spp. [13-18]. These methods have greatly enhanced knowledgeon the epidemiology of oral and subgingivalCandida spp., and can provide valuable informationthrough their ability to distinguish distinctisolates of the same species. Some studies havedemonstrated that commensal yeasts dominate inoral candidiasis, whereas controversial evidenceshows that genetically homogeneous, hypervirulentstrains of C. albicansare involved in the disease [19]. Since there is no available data on the epidemiologyof yeasts and genetic characterization of periimplant C. albicans, the aim of this study was tocharacterize periimplantbiofilm and mucosal C. albicansisolates recovered from immuno competentsubjects with more than 5 years of implant treatment, and to assay the genetic similarityof C. albicans isolates from the two niches in thesame patient by RAPD.
Material and Methods
Study population
This study was approved by the Ethics Committee of the School of Pharmacy and Biochemistry, University of Buenos Aires (Res. 41, File 727.071/10). Yeasts recovered from periimplant plaque (n = 242) and buccal samples (n = 300) were studied in 100 immunocompetent nonsmoking patients with more than five years of implant treatment on oral prosthesis who attended the dental clinic of the Asociacion Implantodontologica Argentina, Buenos Aires, Argentina. Evaluations included clinical examination and radiographs with clinical measurements: pocket depth (PD), considered regular up to 3 mm around implants, plaque index, gingival index [11,20] and bleeding on probing. Measurements were taken at four sites per tooth (mesial, buccal, distal and lingual positions) on 15 teeth, excluding third molars.
This study was approved by the Ethics Committee of the School of Pharmacy and Biochemistry, University of Buenos Aires (Res. 41, File 727.071/10). Yeasts recovered from periimplant plaque (n = 242) and buccal samples (n = 300) were studied in 100 immunocompetent nonsmoking patients with more than five years of implant treatment on oral prosthesis who attended the dental clinic of the Asociacion Implantodontologica Argentina, Buenos Aires, Argentina. Evaluations included clinical examination and radiographs with clinical measurements: pocket depth (PD), considered regular up to 3 mm around implants, plaque index, gingival index [11,20] and bleeding on probing. Measurements were taken at four sites per tooth (mesial, buccal, distal and lingual positions) on 15 teeth, excluding third molars.
Bone resorption was assessed by comparing the radiographic examination in the patients’ medical records taken at the time of implant placement to those taken at the appointment for this study. In order to analyze bone resorption, implants were classified into two groups according to time of implant placement: “immediately loaded implants” if they were placed during the same session as tooth extraction or “delayed loaded implants” if they were placed on healed bone, months or years after extraction. Participation in our survey was voluntary and all patients provided written informed consent. The volunteers were requested to rinse their mouths thoroughly with sterile distilled water, after which sterile swabs were used to take samples from tongue, palate and cheek.
The dental professional then isolated the area using cotton rolls and a highspeed suction device. Following removal of the supragingival plaque using a Teflon curette to avoid salivary contamination, periimplant biofilm was collected from the interdental plate by inserting 34 sterile paper points number 30-35-40 for 15-30 minutes in the four sites: mesial, buccal, distal and lingual positions. Samples were cultured in a differential chromogenic médium (CHROMagar Candida, Paris, France). Yeast isolates were identified using conventional mycological methods: colony color on the chromogenic medium, micromorphology in agar milk with 1% Tween80 [21], carbohydrate assimilation tests using a commercially available kit API ID 32D (BioMerieux, Lyon, France), and specific PCR [22].
Random amplified polymorphic DNA (RAPD) analysis
Yeast DNA was isolated using a technique described previously [22-24]. Five different primers were included in the typing assays. Primer sequences were asfollows: OPA 02 (TGCCGAGCTG), OPA 09 (GGGTAACGCC), M13F (CGACGTTGTAAAACGACGCCCAGT), M13R (CAGGAAACAGCTATGAC), and OCP 5 (GATGACCGCC). They were all used in RAPDPCR, following the method developed by Williams., et al. [23] Arbitrary amplification was performed in a total volume of 50 μl containing: 1_buffer 2.5 mM MgCl2, 0.2 mM each of the dNTP, 0.5 mM of the primer, 1.25 U Taq DNA polymerase (Invitrogen), and 75 ng of template DNA. The cycling program consisted of 4 min at 94°C, 35 1minute cycles at 94°C, 1 min at 25°C, 2 min at 72°C followed by a final extension of 5 min at 72°C. These steps were carried out in a Minicycler DNA thermal cycler (TM MJ Research Inc., NY, USA).
Yeast DNA was isolated using a technique described previously [22-24]. Five different primers were included in the typing assays. Primer sequences were asfollows: OPA 02 (TGCCGAGCTG), OPA 09 (GGGTAACGCC), M13F (CGACGTTGTAAAACGACGCCCAGT), M13R (CAGGAAACAGCTATGAC), and OCP 5 (GATGACCGCC). They were all used in RAPDPCR, following the method developed by Williams., et al. [23] Arbitrary amplification was performed in a total volume of 50 μl containing: 1_buffer 2.5 mM MgCl2, 0.2 mM each of the dNTP, 0.5 mM of the primer, 1.25 U Taq DNA polymerase (Invitrogen), and 75 ng of template DNA. The cycling program consisted of 4 min at 94°C, 35 1minute cycles at 94°C, 1 min at 25°C, 2 min at 72°C followed by a final extension of 5 min at 72°C. These steps were carried out in a Minicycler DNA thermal cycler (TM MJ Research Inc., NY, USA).
Products were separated by electrophoresis in 1.4% agarose gel and stained with ethidium bromide. They were visualized under UV light and digitalized by image analyzer software (EPIChemi Darkroom. UVP Laboratory Products, California, USA). Band profiles were analyzed and compared visually. Each band was scored as positive or negative for all isolates and the presence or absence of each bandwas recorded for each isolate. The resulting matrix was interpreted using the Treecon program, where isolates were grouped according to the resemblance of their patterns. Based on matrix of similarity co-efficients (SC), a dendrogram was generated by the unweighted pair group method using arithmetic averages (UPGMA). The criterion used for genotyping was as follows: arbitrary threshold at an SC of 90% for closely related isolates.
Statistical analysis
Statistical analysis was performed using Statistix 7.0 and the SPSS 11.0 software. Confidence intervalwas 95% (CI 95%). Fisher and ANOVA were calculated at 95% using the EpiInfo 6.04 program (Atlanta University, GA).
Statistical analysis was performed using Statistix 7.0 and the SPSS 11.0 software. Confidence intervalwas 95% (CI 95%). Fisher and ANOVA were calculated at 95% using the EpiInfo 6.04 program (Atlanta University, GA).
Results and Discussion
Clinical features
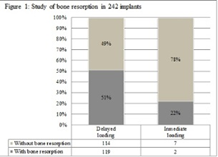
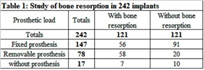
The 100 subjects included in the study ranged in age from 25 to 94 years (mean age 60.1 years), 65% were female (65/100). None of them had received antibacterial or antifungal agents before this treatment. Of the total population, 75% were nonsmokers. This population had an average of 15.34 teeth and 2.56 implants; 2.25 loaded implants and 0.17 nonloaded implants. Of the total number of original implants (n = 256) in the study population, we found that only 242 were present. The percentage of bone resorption in immediately loaded implants (n = 25), was significantly lower (p < 0.001) than in delayed loaded implants (n = 217) (Figure 1). Comparison of bone resorption in relation to the kind of prosthesis placed on the implants (n = 242) showed significantly higher resorption rates (p < 0.001) in the group with removable prostheses (49/78) than in the group with fixed prostheses (34/147) and without load (8/17) (Table 1). Pocket depth (PD) was more than 3 mm in 34/242 samples and less than 3 mm in 208/242 samples (Table 2).
The 100 subjects included in the study ranged in age from 25 to 94 years (mean age 60.1 years), 65% were female (65/100). None of them had received antibacterial or antifungal agents before this treatment. Of the total population, 75% were nonsmokers. This population had an average of 15.34 teeth and 2.56 implants; 2.25 loaded implants and 0.17 nonloaded implants. Of the total number of original implants (n = 256) in the study population, we found that only 242 were present. The percentage of bone resorption in immediately loaded implants (n = 25), was significantly lower (p < 0.001) than in delayed loaded implants (n = 217) (Figure 1). Comparison of bone resorption in relation to the kind of prosthesis placed on the implants (n = 242) showed significantly higher resorption rates (p < 0.001) in the group with removable prostheses (49/78) than in the group with fixed prostheses (34/147) and without load (8/17) (Table 1). Pocket depth (PD) was more than 3 mm in 34/242 samples and less than 3 mm in 208/242 samples (Table 2).
Carriage of C. albicans and other yeast species
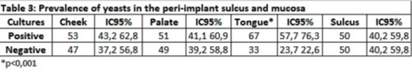
The prevalence of yeasts in the periimplant sulcus was 37.6% (n = 91/242, CI 95%: 31.5 43.7). In buccal mucosa, the distribution of yeasts was: 51% in palate (51/100 patients, CI 95% 41.2-60.9), 53% in cheek (53/100, CI95%: 43.2-62.9) and 67% in lingual mucosa (67/100, CI 95%57.7-76.3), representing a high statistically significant prevalence (p < 0.001) (Table 3).
The prevalence of yeasts in the periimplant sulcus was 37.6% (n = 91/242, CI 95%: 31.5 43.7). In buccal mucosa, the distribution of yeasts was: 51% in palate (51/100 patients, CI 95% 41.2-60.9), 53% in cheek (53/100, CI95%: 43.2-62.9) and 67% in lingual mucosa (67/100, CI 95%57.7-76.3), representing a high statistically significant prevalence (p < 0.001) (Table 3).
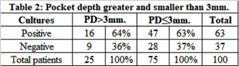
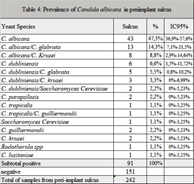
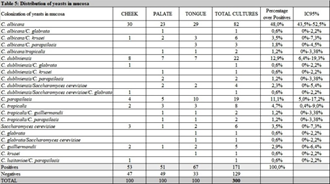
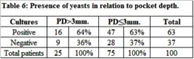
Table 4 summarizes species distribution of yeast isolates in periimplant biofilm and buccal mucosa. Of the 262 yeasts recovered in mucosa (n = 171) and periimplant (n = 91), C. albicans was the species most frequently found in all niches. The prevalence of C. albicans was 47.3% (n = 43/91) in periimplant biofilm. Other nonC. albicans spp.and other yeasts were found: C. dubliniensis (n = 13), C. parapsilosis (n = 13), C. tropicalis (n = 7), Saccharomyces cereviciae (n = 6), C. C. krusei (n = 5), guilliermondii (n = 3), C. glabrata (n = 3), Rhodotorula spp. (n = 2) and C. lusitaniae (n = 1). The occurrence of two or three isolatedspecieswas observed in 27/171 positives buccal mucosa samples. C. albicans and C. krusei (n = 6) followed by Saccharomyces cereviciaeand C. dubliniensis (n = 4) were the associations most frequentlyobserved. The combinations in periimplantsulcus was 8.79 % (n = 8/91). Of the associations of the species found, themost predominant were C. dubliniensis with C. krusei, and C. albicans with C. glabrata (2% each) (Table 5). In relation to pocket depth and presence of yeasts, patients with periimplantsulcus > 3 mm exhibitedan increase in positive cultures (64%, 16/25) compared to negative cultures (36%, 9/25), where as patients with periimplantsulcus ≤ 3 mm, positive cultures (63%, 47/75) and negativecultures (37%, 28/75) exhibited much lowerdiscrepancy. This difference was not statisticallysignificant (Table 6).
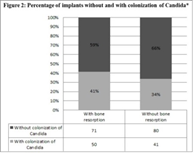
Of the 242 implants studied, 151 showed nocolonization by Candida, of which 71 had bone resorption (47 %) and 80 did not (53%). Of the 91implants where there was colonization by Candida, 50 had resorption (55%) while the other 41 did not (45%). In all four cases, the percentages weresimilar. According to these results, periimplant Candida colonization would not be the determiningcause of bone resorption around implants. (Figure 2)
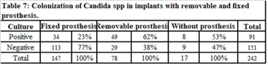
Implants with removable prostheses exhibitedsignificantly higher (p < 0.001) rates of Candida spp. colonization63 %( 49/78) than those with fixedprostheses (34/147) (Table 7).
Rapd-Pcr Assay
We selected five RAPD primers, based on their reproducibility, after the prescreening Process in order to analyze 41C. Albicans isolates. The number of bands ranged from two to three splitters (M13r) to 12 (M13f). Three of five primers were the most informative (M13f, OPA 9 and OPC5) and generated the highest number of band patterns (10 to 12).
We selected five RAPD primers, based on their reproducibility, after the prescreening Process in order to analyze 41C. Albicans isolates. The number of bands ranged from two to three splitters (M13r) to 12 (M13f). Three of five primers were the most informative (M13f, OPA 9 and OPC5) and generated the highest number of band patterns (10 to 12).
The dendrogram generated by the UPGMA clustering method, using the RAPD-PCR technique for C. albicans in oral cavity, tongue (LE), palate (PA), cheek (CA), and periimplant sulcus (I) shows similarity co-efficient (SC) ranging from 60% to 100%. Thirteen genetic clusters and nine main genotypes were obtained at a similarity co-efficient (SC) of 90%. They weredenominated from group 1 to group 9. (Figure 3)
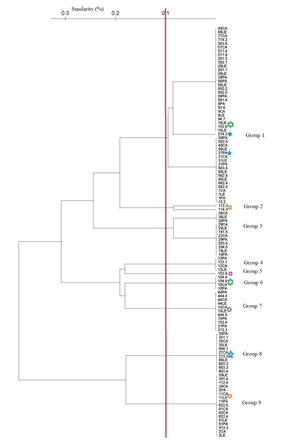
Figure 3: The dendrogram generated by the UPGMA clustering method, using the coefficient of similarity
between RAPD-PCR of C. albicans in oral cavity, tongue (LE), palate (PA), cheek (CA), and periimplant
sulcus (I) shows that the similarity coefficient (SC) ranged from 60 to 100%. Thirteen genetic clusters and
nine main genotypes were obtained at a similarity coefficient (SC) of 90%, genotypes 1, 2, 3, 4 and 5.
Discussion
In this study, 100 immunocompetent adult patients with more than 5 years’ treatment were recruited and grouped according to their health status and pocket depth into periimplantitis or healthy. As expected, patients with periimplantitis presented more infectious sites, including higher rates of percentage similarity (PS) (Anova Test p < 0.001). Eightynine periimplant sulcus samples and 300 swabs from buccal mucosa were cultured directly in CHROMagar Candida medium to enable the presumptive identification of C. albicans or C. dubliniensis, C. tropicalis and C. krusei. This also enabled identification of the presence of infections caused by more than one species simultaneously. Similar findings have been reported by other authors who analyzed other populations [22, 25-28].
The prevalence of yeasts in sulcus was 37.6% (n = 91/242, CI 95%: 31.5 43.7), showing that the surrounding ecological niche and periimplant sulcus enabled yeast growth. Other studies have reported the presence of Candida spp. in periimplant lesions [29,30] and found Candida spp. in 55% of periimplant sites. The comparison of yeast distribution in relation to clinical markers of periimplantitis revealed no significant difference in the prevalence of yeasts at sites with PD >3 mm or at sites with bone resorption.
These findings revealed the presence of yeast species in periimplant sulcus as well at sites with or without periimplantitis. Of the 300 buccal mucosa samples studied here, the tongue was the site with highest prevalence of Candida spp. (67, CI 95% 57.7-76.3), in contrast to cheek and palate, with a statistically significant difference (p < 0.001). Candida spp. prevalence was higher in our study tan in previously reported series [31-34] in which it ranged from 25% to 65%, suggesting that the presence of implants in our study population increases prevalence. In relation to the type of implant rehabilitation–fixedor removable−the latter yielded significantly higher (p < 0.001) prevalence of yeasts. It is worth noting that these findings suggest that periimplant plaque is an ecological niche that favors the growth of yeast species;especially in implants with removable rehabilitation, even though they can be removed for cleaning. Moreover, these implants are made of acrylic, which favors adhesion of Candida spp. These are the first data results reported in Argentina.The use of buccal devices induces alterations within the oral cavity.
Hagg., et al. [35] observed that the presence of prosthesis or other buccal devices increases the number of Candida spp., not only at the site but throughout the mucosa. Dental prostheses are made of acrylic resins in which surface defects favor the development of plaque and prevent its removal [36]. The surface of the prosthesis is very porous and thus susceptible to being colonized by large numbers of microorganisms, which may give rise to different pathologies in the oral cavity.
Comparison of the two study samples showed “high” concordance, with colonization or infection by the same yeast in both ecological niches in 95% of the patients (Kappa = 0.8). In relation to the distribution of yeast species, C. albicans spp. wasthe most prevalent47.3% (n=43/91), but it is important to highlight that nonC.Albicansspp. Were also found in periimplant sulcus: C. dubliniensis C. Dubliniensis 14.3% (n = 13), C. parapsilosis 14.3 %( n = 13), C. tropicalis 7.7% (n = 7), Saccharomyces cereviciae6.6% (n = 6), C. C. krusei 5, 5% (n = 5), guilliermondii 3.3 (n = 3), C. glabrata 3.3% (n = 3), Rhodotorula spp. 2, 2% (n = 2) andC. Lusitaniae 1 %( n = 1). (Table 1). Many of these less prevalent species are emerging and characterized by the presence of diminished sensitivity to antifungals [37]. No data is available in the literature reviewed.
Epidemiological surveillance is very important for identifying the prevalence of yeast species in the biofilm of periimplant sulcus since them créate reservoirs for opportunistic microorganisms which, in certain clinical situations such as patients with immune deficiencies, play a significant role in diseases such as buccal candidiasis and disseminated diseases [34, 38]. In this study, C. albicans isolates from the buccal cavity and periimplant sulcus of the same patient were considered to be closely related in 90% of the cases (27/31) according to RAPD-PCR. Similarity among isolates from both ecological niches suggests that the source of C. albicans colonization in periimplantbiofilm is the patient’s buccal cavity.
Thus, it can be assumed that most C. albicans spp. found in periimplant biofilm originate from endogenous infection by commensal strains. Coincidentally, other authors have found identical genetic patterns in yeasts from different anatomical sites in the same patient. However, the results obtained highlight the fact that the same patient carries different species [39]. It is important to consider that C. albicanscolonization in periimplant sulcus could also occur due to the presence strains adaptable to the periimplant environment, which is likely as a result of genetic variations such as gene conversion and/or chromosomal translocations [15,19]. To date, scientific literature has not provided any information on the genetic characterization of C. albicansisolates in periimplant sulcus. Hence, yeast isolates were analyzed by RAPD-PCR, which has proved to be a rapid, simple, costeffective technique and discriminatory for the molecular typing of C. albicansisolates. Other authors have used the same techniques to assay several yeasts species [13, 15-17, 22].
This is the first study conducted in Argentina on the molecular characterization of clinical C. albicans isolates in periimplant sulcus by RAPDPCR. We confirm that the periimplant plaque is anecological niche that favors the growth of yeast species; Especially in implants with removable rehabilitation. C. albicans spp. were the most prevalent in periimplant samples, but it is important to highlight that non C. albicans spp. were also found in periimplant sulcus, e.g. C. dubliniensis, C. parapsilosis, Saccaromyces cereviciae, C. tropicalis, C. lusitaniaeand C. krusei. The findings suggest that most periimplant C. Albicansoriginate from endogenous infection bycommensal strains.
References
- Mombelli A. “Aging and the periodontal and periimplant microbiota”. Periodontol 2000 16 (1998): 44-52.
- Marsh PD. “Dental plaque: biological significance of a biofilm and community lifestyle”. Journal of Clinical Periodontology 32.6 (2005): 7-15.
- Listgarten MA., et al. “Microbial composition and pattern of antibiotic resistance in subgingival microbial samples from patients with refractory periodontitis”. Journal of Clinical Periodontology 64.3 (1993): 155-161.
- Marsh PD. “Plaque as a biofilm: pharmacological principles of drug delivery and action in the suband supragingival environment”. Oral Diseases 9.1 (2003): 16-22.
- Pye AD., et al. “A review of dental implants and infection”. Journal of Hospital Infection 72.2 (2009): 104-110.
- Maximo MB., et al. “Shortter clinical and microbiological evaluations of periimplant diseases before and after mechanical antiinfective therapies”. Clinical Oral Implants Research 20.1 (2009): 99-108.
- Norowski PA Jr and Bumgardner JD. “Biomaterial and antibiotic strategies for periimplantitis: a review”. Journal of Biomedical Materials Research 88.2 (2009): 530-543.
- Jarvensivu A., et al. “Candida yeasts in chronic periodontitis tissues and subgingival microbial biofilms in vivo”. Oral Diseases 10.2 (2004): 106-112.
- Dahlen G and Wikstrom M. “Occurrence of enteric rods, staphylococci and Candida in subgingival samples”. Oral Microbiology and Immunology 10.1 (1995): 42-46.
- Reynaud AH., et al. “Yeasts in periodontal pockets”. Journal of Clinical Periodontology 28.9 (2001): 860-864.
- Pizzo G., et al. “Biotypes and randomly amplified polymorphic DNA (RAPD) profiles of subgingival Candida albicans isolates in HIV infection”. New Microbiologica 28.1 (2005): 75-82.
- Brady LJ., et al. “Oral diseases, mycology and periodontal microbiology of HIV1infected women”. Oral Microbiology and Immunology 11.6 (1996): 371-380.
- Jain P., et al. “Variation in random amplified polymorphic DNA (RAPD) profiles specific to fluconazoleresistant and sensitive strains of Candida albicans”. Diagnostic Microbiology and Infectious Disease 41.3 (2001): 113-119.
- Dassanayake RS and Samaranayake LP. “Amplificationbased nucleic acid scanning techniques to assess genetic polymorphism in Candida”. Critical Reviews in Microbiology 29.1 (2003): 1-24.
- Montour L., et al. “solation of Candida dubliniensis in an aboriginal community in Ontario, Canada”. Journal of Clinical Microbiology 41.7 (2003): 3423-3426.
- Samaranayake YH., et al. “Genotypic shuffling’ of sequential clones of Candida albicans in HIVinfected individuals with and without symptomatic oral candidiasis”. Journal of Medical Microbiology 52.4 (2003): 349-359.
- Costa F., et al. “Genotypic analysis of Candida albicans isolates obtained from removable prosthesis wearers”. Letters in Applied Microbiology 46.4 (2008): 445-449.
- Jewtuchowicz VM., et al. “Genetic relatedness of subgingival and buccal Candida dubliniensis isolates in immunocompetent subjects assessed by RAPD-PCR”. Journal of Oral Microbiology (2009).
- Song X., et al. “Genetic relatedness of oral yeasts within and between patients with marginal periodontitis and subjects with oral health”. Journal of Periodontal Research 40.6 (2005): 446-452.
- Silness J and Loe H. “Periodontal Disease in Pregnancy. Correlation between Oral Hygiene and Periodontal Condition”. Acta Odontologica Scandinavica (1964): 121-135.
- Jitsurong S., et al. “New milk medium for germ tube and chlamydoconidia production by Candida albicans”. Mycopathologia 123.2 (1993): 95-98.
- Jewtuchowicz VM., et al. “Phenotypic and genotypic identification of Candida dubliniensisfrom subgingival sites in immunocompetent subjects in Argentina”. Oral Microbiology and Immunology 23.6 (2008): 505-509.
- Scherer S and Stevens DA. “Application of DNA typing methods to epidemiology and taxonomy of Candidaspecies”. Journal of Clinical Microbiology 25.4 (1987): 675-679.
- Duran EL., et al. “Examination of the genetic variability among biofilmforming Candida albicansclinical isolates”. Revista Iberoamericana de Micología 24.4 (1987): 268-271.
- Williams JG., et al. “DNA polymorphisms amplified by arbitrary primers are useful as genetic markers”. Nucleic Acids Research 18.22 (1990): 6531-6535.
- Soll DR. “The ins and outs of DNA fingerprinting the infectious fungi”. Clinical Microbiology Reviews 13.2 (2000): 332-370.
- Mujica MT., et al. “Prevalence of Candida albicans and Candida nonalbicansin clinical samples during 1999-2001”. Revista Argentina De Microbiologia 36 (2004): 107-112.
- Jewtuchowicz VM., et al. “Subgingival Subgingival distribution of yeast and their antifungal susceptibility in immunocompetent subjects with and without dental devices”. Acta Odontol Latinoam 20 (2007): 17-22.
- Laine P., et al. Failed dental implants clinical, radiological and bacteriological findings in 17 patients. Journal of Craniofacial Surgery 33.3 (2005): 212-217.
- Salvi GE., et al. “Oneyear bacterial colonization patterns of Staphylococcus aureus and other bacteria at implants and adjacent teeth”. Clinical Oral Implants Research 19.3 (2008): 242-248.
- Kleinegger CL., et al. “Frequency, intensity, species, and strains of oral Candida vary as a function of host age”. Journal of Clinical Microbiology 34.9 (1996): 2246-2254.
- Aguirre Urizar JM. “Oral candidiasis”. Revista Iberoamericana de Micologí 19 (2002): 17-21.
- Negroni M., et al. “Candida carriage in the oral mucosa of a student population: adhesiveness of the strains and predisposing factors”. Revista Argentina De Microbiologia 34.1 (2002): 22-28.
- Luque AG., et al. “Oral yeast carriage in HIV infected and non-infected populations in Rosario, Argentina”. Mycoses 52.1 (2009): 53-59.
- Hagg U., et al. “The effect of fixed orthodontic appliances on the oral carriage of Candida species and Enterobacteriaceae”. European Journal of Orthodontics 26.6 (2004): 623-629.
- Verran J and Maryan CJ. “Retention of Candida albicans on acrylic resin and silicone of different surface topography”. Journal of Prosthetic Dentistry 77.5 (1997): 535-539.
- Hazen KC. “New and emerging yeast pathogens”. Clinical Microbiology Reviews 8.4 (1995): 462-478.
- Gonzalez Gravina H., et al. “Oral Candidiasis in children and adolescents with cancer. Identification of Candida spp”. Medicina Oral, Patologia Oral Y Cirugia Bucal 12.6 (2007): 419-423.
- Badoc C., et al. “Clonality structure in Candida dubliniensis”. FEMS Microbiology Letters 209.2 (2002): 249-254.
Citation:
Virginia M Jewtuchowicz., et al. “Candida albicans RAPD-PCR Genotypic Study in Peri-implant Sulcus and Buccal Mucosa”.
Oral Health and Dentistry 2.1 (2017): 312-320.
Copyright: © 2017 Virginia M Jewtuchowicz., et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.



































 Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.
Scientia Ricerca is licensed and content of this site is available under a Creative Commons Attribution 4.0 International License.